Association Between Klotho Gene Polymorphisms and Urolithiasis: A Meta-Analysis
By Jiaxuan Qin, Bo Duan, Bowen Chen, Jinchun Xing, Tao WangAffiliations
doi: 10.29271/jcpsp.2024.02.206ABSTRACT
Former studies have suggested that urolithiasis is related to Klotho gene polymorphisms. The aim of this meta-analysis was to investigate this relationship. Studies on the association between urolithiasis susceptibility and Klotho gene polymorphisms were systematically searched for in databases. Odds ratios and 95% confidence intervals were pooled as the effect size. This meta-analysis incorporated ten articles. Klotho rs1207568 adenine (A) may be related to a decreased urolithiasis risk in Caucasians. The results showed that Klotho rs3752472 may not be related to urolithiasis risk in the Han Asian subgroup. Klotho rs564481 may not be related to urolithiasis risk in Asians or Caucasians, and Klotho rs650439 may not be related to urolithiasis risk in Asians.
Key Words: Klotho, Single-nucleotide polymorphism, Urolithiasis, Meta-analysis.
INTRODUCTION
Urolithiasis is likely related to multiple genes’ effects, and related to environmental and lifestyle factors.1,2 Calcium stone is the major type of urolithiasis. Infection can promote the stone formation. Klotho might be associated with inflammatory processes in kidney injury.3,4 The Klotho protein, a Type-I transmembrane protein expressed by the Klotho gene on chromosome 13q13.1 in tissues responsible for calcium homeostasis, including the kidney and epithelium of the choroid plexus in the brain and parathyroid gland, plays a crucial role in phosphate homeostasis regulation via FGF23 and increased calcium uptake via TRPV5.5,6 The single nucleotide polymorphism (SNP) rs1207568 (G395A) is located in Klotho gene’s promoter region, and DNA-protein interaction can be affected by the G-A substitution in this region.7 Calcium oxalate crystals can cause renal epithelial cell injury, which may be prevented by Klotho rs3752472.8 Previous studies in several populations have suggested that urolithiasis may be associated with Klotho polymorphisms. Here, this meta-analysis was conducted to evaluate it, and provide specific genetic markers for urolithiasis.
METHODOLOGY
In EMBASE, China National Knowledge Infrastructure (CNKI), clinicaltrials.gov, PubMed, and Cochrane Library databases, two independent investigators performed a systematic search on 23 April 2022.
The following terms were used without any limitations: “Klotho or HFTC3” and “polymorphisms or polymorphism” and “calculi or stone or nephrolithiasis or calculus or urolithiasis or lithiasis.” The references of related reviews and studies were artificially indexed.
Examples were taken from meta-analysis published by the team to establish the inclusion and exclusion criteria.9,10 It was attempted to email the author for detailed genotype data. Two investigators independently performed the study selection. Any disputes were resolved through discussions. If necessary, another investigator may be invited to participate in further discussions. Detective samples, year of publication, Hardy-Weinberg equilibrium, first author’s surname, urinary calculi’s chemical composition, genotyping method, ethnicity, source of control groups, characteristics, country of origin, and number for each genotype were collected.
Independently, two investigators evaluated absorbed studies’ quality by using Newcastle-Ottawa Scale (NOS).11 The most important factors were country, gender, age, and ethnicity. The second important factor was urolithiasis-related diseases that alter calcium or phosphorus metabolism, such as hyperparathyroidism and a family history of urolithiasis. The quality scores ranged from 0 to 10. Detailed statistical analysis was conducted as relevant.12-14
RESULTS
Finally, 61 articles were identified from the databases (EMBASE = 29, PubMed = 15, CNKI = 14, clinicaltrials.gov = 2, Cochrane = 1, other sources (manual search) = 0). Figure 1 illustrates the screening process. Nine full-text articles were excluded, three being without detailed genotype data,15-17 and six being duplicate studies. Ultimately, 11 articles were absorbed in this meta-analysis.18-28
Table I: Characteristics of studies.
|
No. |
Study ID |
Year |
Country or area |
Ethnicity |
Control type |
Genotyping method |
Urolithiasis related diseases# in |
Family history of urolithiasis in patients |
Stone composition |
P for HWE* |
Quality |
|
|
rs1207568 (G395A) |
|
|
|
|
|
|
|
|
|
|
|
1.1.1 |
Telci et al. 18 |
2011 |
Turkiye |
Caucasian |
PB* |
PCR-RFLP |
Negative |
NA* |
Calcium stone |
0.006& |
7 |
|
1.1.2 |
Gürel et al. 19 |
2016 |
Turkiye |
Caucasian |
HB* |
PCR-RFLP |
NA |
NA |
NA |
0.000 |
6 |
|
1.1.3 |
Lanka et al. 20 |
2021 |
Northwestern India |
Caucasian |
PB |
PCR-RFLP |
Negative |
NA |
88.7% Calcium oxalate stone; 11.3% Calcium oxalate and phosphate stone mixed |
0.002 |
7 |
|
1.2.1 |
Chen et al. 21 |
2013 |
China |
Asian(Han) |
PB |
TaqMan |
Negative |
negative |
Calcium stone |
0.131 |
9 |
|
1.2.2 |
Enli et al. 22 |
2018 |
China |
Asian(Han) |
PB |
Sequencing |
Negative |
negative |
Calcium stone |
0.105 |
7 |
|
1.3.1 |
Ali et al. 23 |
2017 |
China |
Asian(Uyghur) |
PB |
PCR-RFLP |
Negative |
52.3% positive |
Calcium oxalate stone |
0.003 |
7 |
|
1.3.2 |
Qi et al. 24 |
2019 |
China |
Asian(Uyghur) |
PB |
PCR-RFLP& Sequencing |
Negative |
Negative |
Calcium oxalate stone |
0.946 |
8 |
|
- |
rs3752472 |
- |
- |
- |
- |
- |
- |
- |
- |
- |
- |
|
2.1.1 |
Wei et al. 25 |
2015 |
China |
Asian(Han) |
PB |
TaqMan |
Negative |
NA |
pure or mixed Calcium oxalate stone |
0.311 |
7 |
|
2.1.2 |
Enli20 et al. 22 |
2018 |
China |
Asian(Han) |
PB |
Sequencing |
Negative |
Negative |
Calcium stone |
0.455 |
7 |
|
2.1.3 |
Peili et al. 26 |
2020 |
China |
Asian(Han) |
PB |
SNaPshot |
Negative |
Negative |
Calcium oxalate stone with purity≥65% |
0.766 |
8 |
|
2.2.1 |
Ali et al. 23 |
2017 |
China |
Asian(Uyghur) |
PB |
PCR-RFLP |
Negative |
52.3% Positive |
Calcium oxalate stone |
0.124 |
7 |
|
2.2.2 |
Qi et al. 24 |
2019 |
China |
Asian(Uyghur) |
PB |
PCR-RFLP& Sequencing |
Negative |
Negative |
Calcium oxalate stone |
0.730 |
8 |
|
2.3 |
Litvinova et al. 27 |
2021 |
Russia |
Caucasian |
PB |
Sequencing |
NA |
52% positive |
Calcium oxalate stone |
NA |
7 |
|
- |
rs564481 (C1818T) |
- |
- |
- |
- |
- |
- |
- |
- |
- |
- |
|
3.1.1 |
Telci et al. 18 |
2011 |
Turkiye |
Caucasian |
PB |
PCR-RFLP |
Negative |
NA |
Calcium stone |
0.787 |
7 |
|
3.1.2 |
Gürel et al. 19 |
2016 |
Turkiye |
Caucasian |
HB |
PCR-RFLP |
NA |
NA |
NA |
0.647 |
6 |
|
3.2.1 |
Chen et al. 21 |
2013 |
China |
Asian(Han) |
PB |
TaqMan |
Negative |
negative |
Calcium stone |
0.107 |
9 |
|
3.2.2 |
Peili et al. 26 |
2020 |
China |
Asian(Han) |
PB |
SNaPshot |
Negative |
Negative |
Calcium oxalate stone with purity≥65% |
0.421 |
8 |
|
3.3 |
Qi et al. 24 |
2019 |
China |
Asian(Uyghur) |
PB |
PCR-RFLP& Sequencing |
Negative |
Negative |
Calcium oxalate stone |
0.435 |
8 |
| rs650439 | |||||||||||
|
4.1.1 |
Ali et al. 23 |
2017 |
China |
Asian(Uyghur) |
PB |
PCR-RFLP |
Negative |
52.3% positive |
Calcium oxalate stone |
0.360 |
7 |
|
4.1.2 |
Qi et al. 24 |
2019 |
China |
Asian(Uyghur) |
PB |
PCR-RFLP& Sequencing |
Negative |
negative |
Calcium oxalate stone |
0.413 |
8 |
|
4.2 |
Wei et al. 25 |
2015 |
China |
Asian(Han) |
PB |
TaqMan |
Negative |
NA |
Pure or mixed calcium oxalate stone |
0.359 |
7 |
|
- |
F352V |
- |
- |
- |
- |
- |
- |
- |
- |
- |
- |
|
5.1 |
Telci et al. 18 |
2011 |
Turkiye |
Caucasian |
PB |
PCR-RFLP |
Negative |
NA |
Calcium stone |
0.184 |
7 |
|
5.2 |
Gürel et al. 19 |
2016 |
Turkiye |
Caucasian |
HB |
PCR-RFLP |
NA |
NA |
NA |
0.009 |
6 |
|
- |
rs145682430 |
- |
- |
- |
- |
- |
- |
- |
- |
- |
- |
|
6.1 |
Peili et al. 26 |
2020 |
China |
Asian(Uyghur) |
PB |
SNaPshot |
Negative |
Negative |
Calcium oxalate stone with purity≥65% |
0.725 |
8 |
|
6.2 |
Peili et al. 26 |
2020 |
China |
Asian(Han) |
PB |
SNaPshot |
Negative |
Negative |
calcium oxalate Stone with purity≥65% |
0.519 |
8 |
|
- |
rs139912465 |
- |
- |
- |
- |
- |
- |
- |
- |
- |
- |
|
7 |
Liuya et al. 28 |
2015 |
China |
Asian(Uyghur) |
PB |
PCR-RFLP |
Negative |
52.3% Positive |
Calcium oxalate stone |
1 |
7 |
|
- |
rs577912 |
- |
- |
- |
- |
- |
- |
- |
- |
- |
- |
|
8 |
Wei et al. 25 |
2015 |
China |
Asian(Han) |
PB |
TaqMan |
Negative |
NA |
Pure or mixed calcium oxalate stone |
0.153 |
7 |
|
-- |
rs397703 |
- |
- |
- |
- |
- |
- |
- |
- |
- |
- |
|
9 |
Enli et al. 22 |
2018 |
China |
Asian(Han) |
PB |
Sequencing |
Negative |
Negative |
Calcium stone |
0.544 |
7 |
|
- |
rs648202 |
- |
- |
- |
- |
- |
- |
- |
- |
- |
- |
|
10 |
Qi et al. 24 |
2019 |
China |
Asian(Uyghur) |
PB |
PCR-RFLP& Sequencing |
Negative |
Negative |
Calcium oxalate stone |
0.285 |
8 |
|
- |
rs526906 |
- |
- |
- |
- |
- |
- |
- |
- |
- |
- |
|
11.1 |
Apolikhin et al. 15 |
2015 |
Russia |
Caucasian |
PB |
NA |
NA |
NA |
NA |
NA |
NA |
|
11.2 |
Apolikhin et al. 16 |
2016 |
Russia |
Caucasian |
PB |
NA |
NA |
NA |
NA |
NA |
NA |
|
11.3 |
Apolikhin et al. 17 |
2017 |
Russia |
Caucasian |
PB |
NA |
NA |
NA |
Calcium oxalate stone |
NA |
NA |
|
# Diseases altering calcium and phosphorus metabolism like hyperparathyroidism; * HWE: Hardy–Weinberg equilibrium; PB: Population-based; HB: Hospital-based; NA: Not available; & Results with statistical significant difference were marked as bold. |
|||||||||||
Table I and II show the characteristics and detailed genotype data for each study. PCR–RFLP, TaqMan, SNaPshot, and sequencing were used as genotyping methods. Every studies used blood samples for genotyping. Control group of study no 1.1.1, 1.1.2, 1.1.3, 1.3.1 and 5.2 departed from HWE significantly. Owing to the lack of detailed genotype data, HWE was not evaluated in study no 2.3’s control group.
Table III shows the results of pooled ORs. In a meta-analysis, rs1207568 adenine (A) was related to a decreased urolithiasis risk in dominant model (AG + AA vs. GG), heterozygote comparison (AG vs. GG), and allelic comparison (A vs. G) in the Caucasian subgroup. No statistically significant change in urolithiasis risk was discovered in the other genetic models, groups, or subgroups of rs1207568 (Table III and Figure 2).
In a meta-analysis, rs3752472 adenine (A) was related to a decreased urolithiasis risk in homozygote comparison (AA vs. GG) and recessive model (AA vs. GG + AG) in the Uyghur subgroup. No statistically significant change in urolithiasis risk was discovered in the other genetic models, groups, or subgroups of rs3752472. Heterogeneity in most groups and subgroups of rs3752472 was significant (Table III and Figure 3).
In a meta-analysis, no statistically significant change in urolithiasis risk was discovered in any genetic model or subgroup of rs564481, rs650439, F352V, or rs145682430. In each study included in each SNP, no statistically significant changes were discovered.
rs577912 adenine (A) was related to an increased urolithiasis risk in all genetic models. No statistically significant change in urolithiasis risk was discovered in any of the genetic models of rs139912465, rs397703, and rs648202. Among these four SNPs, only one study was included in each SNP; therefore, meta-analysis could not be operated.
Table II: Detailed genotype data of studies.
|
No. |
Study ID |
Case |
Control |
||||||||||
|
|
rs1207568 |
GG |
GA |
AA |
G |
A |
Total |
GG |
GA |
AA |
G |
A |
Total |
|
1.1.1 |
Telci et al. 18 |
63 |
41 |
4 |
167 |
49 |
108 |
19 |
31 |
1 |
69 |
33 |
51 |
|
1.1.2 |
Gürel et al. 19 |
54 |
45 |
4 |
153 |
53 |
103 |
32 |
68 |
2 |
132 |
72 |
102 |
|
1.1.3 |
Lanka et al. 20 |
108 |
42 |
0 |
258 |
42 |
150 |
52 |
48 |
0 |
152 |
48 |
100 |
|
1.2.1 |
Chen et al. 21 |
208 |
89 |
9 |
505 |
107 |
306 |
167 |
71 |
3 |
405 |
77 |
241 |
|
1.2.2 |
Enli et al. 22 |
345 |
147 |
11 |
837 |
169 |
503 |
371 |
156 |
25 |
898 |
206 |
552 |
|
1.3.1 |
Ali et al. 23 |
74 |
46 |
8 |
194 |
62 |
128 |
61 |
23 |
10 |
145 |
43 |
94 |
|
1.3.2 |
Qi et al. 24 |
273 |
102 |
25 |
648 |
152 |
400 |
241 |
147 |
22 |
629 |
191 |
410 |
|
|
rs3752472 |
GG |
GA |
AA |
G |
A |
Total |
GG |
GA |
AA |
G |
A |
Total |
|
2.1.1 |
Wei et al.25 |
1464 |
220 |
20 |
3148 |
260 |
1704 |
904 |
208 |
16 |
2016 |
240 |
1128 |
|
2.1.2 |
Enli et al. 22 |
406 |
90 |
6 |
902 |
102 |
502 |
457 |
93 |
3 |
1007 |
99 |
553 |
|
2.1.3 |
Peili et al. 26 |
331 |
75 |
6 |
737 |
87 |
412 |
355 |
44 |
1 |
754 |
46 |
400 |
|
2.2.1 |
Ali et al. 23 |
113 |
14 |
1 |
240 |
16 |
128 |
63 |
25 |
6 |
151 |
37 |
94 |
|
2.2.2 |
Qi et al. 24 |
349 |
50 |
0 |
748 |
50 |
399 |
349 |
58 |
3 |
756 |
64 |
410 |
|
2.3 |
Litvinova et al. 27 |
NA* |
NA |
NA |
98 |
2 |
50 |
NA |
NA |
NA |
98 |
2 |
50 |
|
|
rs564481 |
GG |
GA |
AA |
G |
A |
Total |
GG |
GA |
AA |
G |
A |
Total |
|
3.1.1 |
Telci et al. 18 |
47 |
46 |
15 |
140 |
76 |
108 |
19 |
25 |
7 |
63 |
39 |
51 |
|
3.1.2 |
Gürel et al. 19 |
45 |
41 |
17 |
131 |
75 |
103 |
33 |
52 |
17 |
118 |
86 |
102 |
|
3.2.1 |
Chen et al. 21 |
199 |
GA+AA=107 |
data error |
306 |
159 |
78 |
4 |
396 |
86 |
241 |
||
|
3.2.2 |
Peili et al. 26 |
267 |
128 |
17 |
662 |
162 |
412 |
257 |
124 |
19 |
638 |
162 |
400 |
|
3.3 |
Qi et al. 24 |
201 |
164 |
34 |
566 |
232 |
399 |
196 |
170 |
44 |
562 |
258 |
410 |
|
|
rs650439 |
AA |
AT |
TT |
A |
T |
Total |
AA |
AT |
TT |
A |
T |
Total |
|
4.1.1 |
Ali et al. 23 |
81 |
40 |
7 |
202 |
54 |
128 |
56 |
31 |
7 |
143 |
45 |
94 |
|
4.1.2 |
Qi et al. 24 |
213 |
158 |
28 |
584 |
214 |
399 |
217 |
167 |
26 |
601 |
219 |
410 |
|
4.2 |
Wei et al.25 |
812 |
720 |
172 |
2344 |
1064 |
1704 |
508 |
508 |
112 |
1524 |
732 |
1128 |
|
|
F352V |
TT |
TG |
GG |
T |
G |
Total |
TT |
TG |
GG |
T |
G |
Total |
|
5.1 |
Telci et al. 18 |
71 |
33 |
4 |
175 |
41 |
108 |
35 |
16 |
0 |
86 |
16 |
51 |
|
5.2 |
Gürel et al. 19 |
60 |
40 |
3 |
160 |
46 |
103 |
60 |
42 |
0 |
162 |
42 |
102 |
|
|
rs145682430 |
GG |
GA |
AA |
G |
A |
Total |
GG |
GA |
AA |
G |
A |
Total |
|
6.1 |
Peili et al. 26 |
393 |
6 |
0 |
792 |
6 |
399 |
396 |
14 |
0 |
806 |
14 |
410 |
|
6.2 |
Peili et al. 26 |
379 |
33 |
0 |
791 |
33 |
412 |
375 |
25 |
0 |
775 |
25 |
400 |
|
|
rs139912465 |
GG |
GA |
AA |
G |
A |
Total |
GG |
GA |
AA |
G |
A |
Total |
|
7 |
Liuya et al. 28 |
0 |
0 |
128 |
0 |
256 |
128 |
0 |
0 |
94 |
0 |
188 |
94 |
|
|
rs577912 |
CC |
CA |
AA |
C |
A |
Total |
CC |
CA |
AA |
C |
A |
Total |
|
8 |
Wei et al.25 |
976 |
604 |
124 |
2556 |
852 |
1704 |
704 |
364 |
60 |
1772 |
484 |
1128 |
|
|
rs397703 |
GG |
GA |
AA |
G |
A |
Total |
GG |
GA |
AA |
G |
A |
Total |
|
9 |
Enli et al. 22 |
8 |
135 |
359 |
151 |
853 |
502 |
16 |
168 |
368 |
200 |
904 |
552 |
|
|
rs648202 |
GG |
GA |
AA |
G |
A |
Total |
GG |
GA |
AA |
G |
A |
Total |
|
10 |
Qi et al. 24 |
206 |
163 |
30 |
575 |
223 |
399 |
219 |
167 |
24 |
605 |
215 |
410 |
|
|
rs526906 |
OR*(95%CI*) |
A |
B |
Total |
|
|
|
A |
B |
Total |
||
|
11.1 |
Apolikhin et al. 15 |
Non-significant |
NA |
NA |
75 |
|
|
|
NA |
NA |
189 |
||
|
11.2 |
Apolikhin et al. 16 |
Non-significant |
NA |
NA |
43 |
|
|
|
NA |
NA |
189 |
||
|
11.3 |
Apolikhin et al. 17 |
Non-significant |
NA |
NA |
72 |
|
|
|
NA |
NA |
189 |
||
|
*NA: Not available; OR: Odds ratio; CI: Confidence interval. |
|||||||||||||
Table III: Results of pooled OR.
|
|
Number |
A vs. G |
|
AA vs. GG |
|
AG vs. GG |
|
AG+AA vs. GG |
|
AA vs. GG+AG |
|
|
rs1207568 |
(cases/controls) |
OR#(95%CI#) |
I2(%) |
OR(95%CI) |
I2(%) |
OR(95%CI) |
I2(%) |
OR(95%CI) |
I2(%) |
OR(95%CI) |
I2(%) |
|
Overall |
1698/1550 |
0.801(0.664-0.966) & |
50.3 |
0.836(0.577-1.211) |
11.7 |
0.701(0.492-0.997) |
78.5 |
0.709(0.521-0.965) |
73.6 |
0.906(0.630-1.303) |
39.4 |
|
Caucasian |
361/253 |
0.587(0.449-0.767) |
0.0 |
1.193(0.299-4.755) |
0.0 |
0.405(0.289-0.570) |
0.0 |
0.420(0.299-0.588) |
0.0 |
1.981(0.508-7.731) |
0.0 |
|
Asian |
1337/1297 |
0.898(0.782-1.031) |
23.8 |
0.812(0.552-1.195) |
44.3 |
0.955(0.675-1.351) |
72.7 |
0.925(0.708-1.208) |
58.8 |
0.854(0.451-1.616) |
56.1 |
|
Han* |
809/793 |
0.951(0.792-1.143) |
28.4 |
0.968(0.198-4.724) |
77.7 |
1.011(0.813-1.257) |
0.0 |
0.979(0.793-1.209) |
0.0 |
0.966(0.198-4.724) |
77.9 |
|
Uyghur* |
528/504 |
0.833(0.675-1.028) |
40.1 |
0.896(0.538-1.493) |
0.0 |
0.970(0.368-2.552) |
87.8 |
0.909(0.455-1.813) |
80.1 |
0.962(0.583-1.588) |
38.9 |
|
rs3752472 |
|
A v.s G |
|
AA vs. GG |
|
AG vs. GG |
|
AG+AA vs. GG |
|
AA vs. GG+AG |
|
|
Overall |
3195/2635 |
0.843(0.537-1.323) |
87.7 |
NA# |
NA |
NA |
NA |
NA |
NA |
NA |
NA |
|
Asian |
3145/2585 |
0.836(0.523-1.334) |
90.2 |
0.828(0.249-2.755) |
63.5 |
0.853(0.548-1.328) |
86.6 |
0.834(0.518-1.343) |
89.0 |
0.874(0.283-2.703) |
58.8 |
|
Han |
2618/2081 |
1.136(0.636-2.028) |
92.5 |
1.653(0.512-5.341) |
58.9 |
1.070(0.597-1.920) |
91.1 |
1.108(0.603-2.035) |
92.1 |
1.600(0.547-4.678) |
52.0 |
|
Uyghur* |
527/504 |
0.477(0.168-1.354) |
87.8 |
0.109(0.019-0.623) |
0.0 |
0.543(0.202-1.464) |
82.6 |
0.488(0.165-1.446) |
86.6 |
0.126(0.022-0.713) |
0.0 |
|
rs564481 |
|
A vs. G |
|
AA vs. GG |
|
AG vs. GG |
|
AG+AA vs. GG |
|
AA vs. GG+AG |
|
|
Overall |
1328/1204 |
0.900(0.781-1.037) |
0.0 |
0.786(0.562-1.101) |
0.0 |
0.901(0.745-1.090) |
0.0 |
0.916(0.779-1.076) |
0.0 |
0.857(0.622-1.181) |
0.0 |
|
Caucasian* |
211/153 |
0.821(0.603-1.116) |
0.0 |
0.780(0.412-1.477) |
0.0 |
0.642(0.403-1.022) |
0.0 |
0.676(0.436-1.046) |
0.0 |
0.998(0.556-1.791) |
0.0 |
|
Asian* |
1117/1051 |
0.923(0.786-1.083) |
0.0 |
0.789(0.531-1.172) |
0.0 |
0.966(0.784-1.191) |
0.0 |
0.961(0.808-1.143) |
0.0 |
0.803(0.546-1.180) |
0.0 |
|
Han* |
718/641 |
NA |
NA |
NA |
NA |
NA |
NA |
1.002(0.801-1.253) |
0.0 |
NA |
NA |
|
rs650439 |
|
T vs. A |
|
TT vs. AA |
|
AT vs. AA |
|
AT+TT vs. AA |
|
TT vs. AA+AT |
|
|
Asian |
2231/1632 |
0.952(0.862-1.051) |
0.0 |
0.968(0.767-1.222) |
0.0 |
0.903(0.789-1.034) |
0.0 |
0.915(0.804-1.040) |
0.0 |
1.019(0.815-1.274) |
0.0 |
|
Uyghur* |
527/504 |
0.973(0.799-1.186) |
0.0 |
0.997(0.603-1.649) |
0.0 |
0.949(0.733-1.229) |
0.0 |
0.955(0.746-1.222) |
0.0 |
1.019(0.623-1.666) |
0.0 |
|
F352V |
|
G vs. T |
|
GG vs. TT |
|
GT vs. TT |
|
GT+GG vs. TT |
|
GG vs. TT+GT |
|
|
Caucasian* |
211/153 |
1.161(0.796-1.694) |
0.0 |
5.560(0.680-45.48) |
0.0 |
0.976(0.627-1.520) |
0.0 |
1.067(0.688-1.652) |
0.0 |
5.592(0.688-45.48) |
0.0 |
|
rs145682430 |
|
A vs. G |
|
AA vs. GG |
|
AG vs. GG |
|
AG+AA vs. GG |
|
AA vs. GG+AG |
|
|
Asian* |
811/810 |
0.811(0.282-2.331) |
73.5 |
NA |
NA |
0.810(0.276-2.374) |
74.0 |
0.810(0.276-2.374) |
74.0 |
NA |
NA |
|
#OR: Odds ratio; CI: Confidence interval; NA: Not available. &Results with statistical significant difference were marked as bold. Unstable results in sensitivity analyses were marked as italic. |
|||||||||||
A sensitivity analysis was performed if any subgroup and any comparison included more than two studies, in dominant model (AG+AA vs. GG), heterozygote comparison (AG vs. GG) and allelic comparison (A vs. G) of rs1207568 overall, statistically different results were gained when study no 1.1.1, 1.1.2, 1.1.3 or 1.3.2 were excluded. In the Asian subgroup of rs1207568, statistically different results were gained when study no 1.2.1 was excluded in allelic comparison (A vs. G, Table III and Figure 2). Less than three studies were included in most groups and subgroups marked with asterisks in Table III; therefore, the sensitivity analysis could not be made. The other results were stable in the sensitivity analysis (Table III).
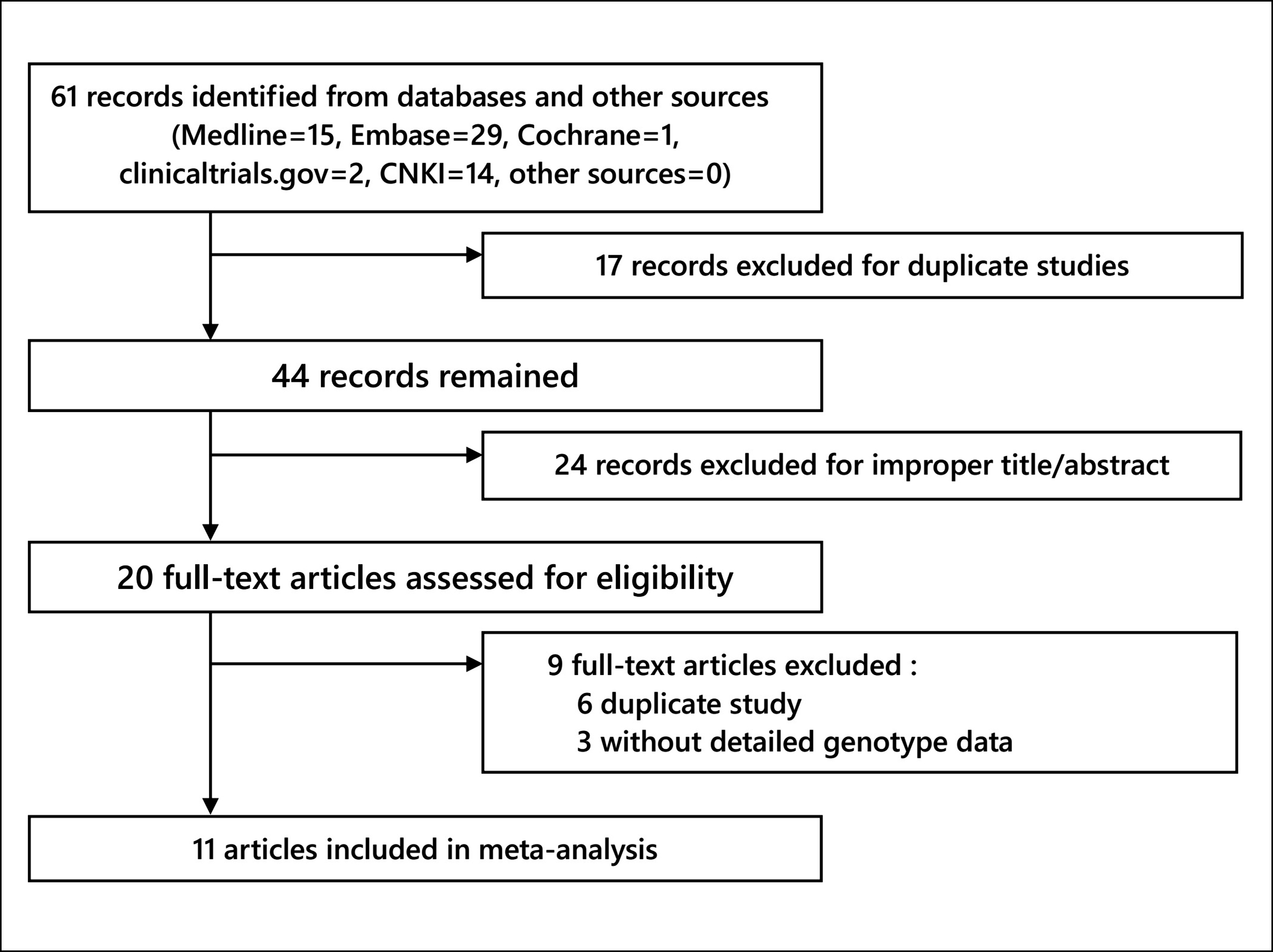 Figure 1. Literature screening process.
Figure 1. Literature screening process.
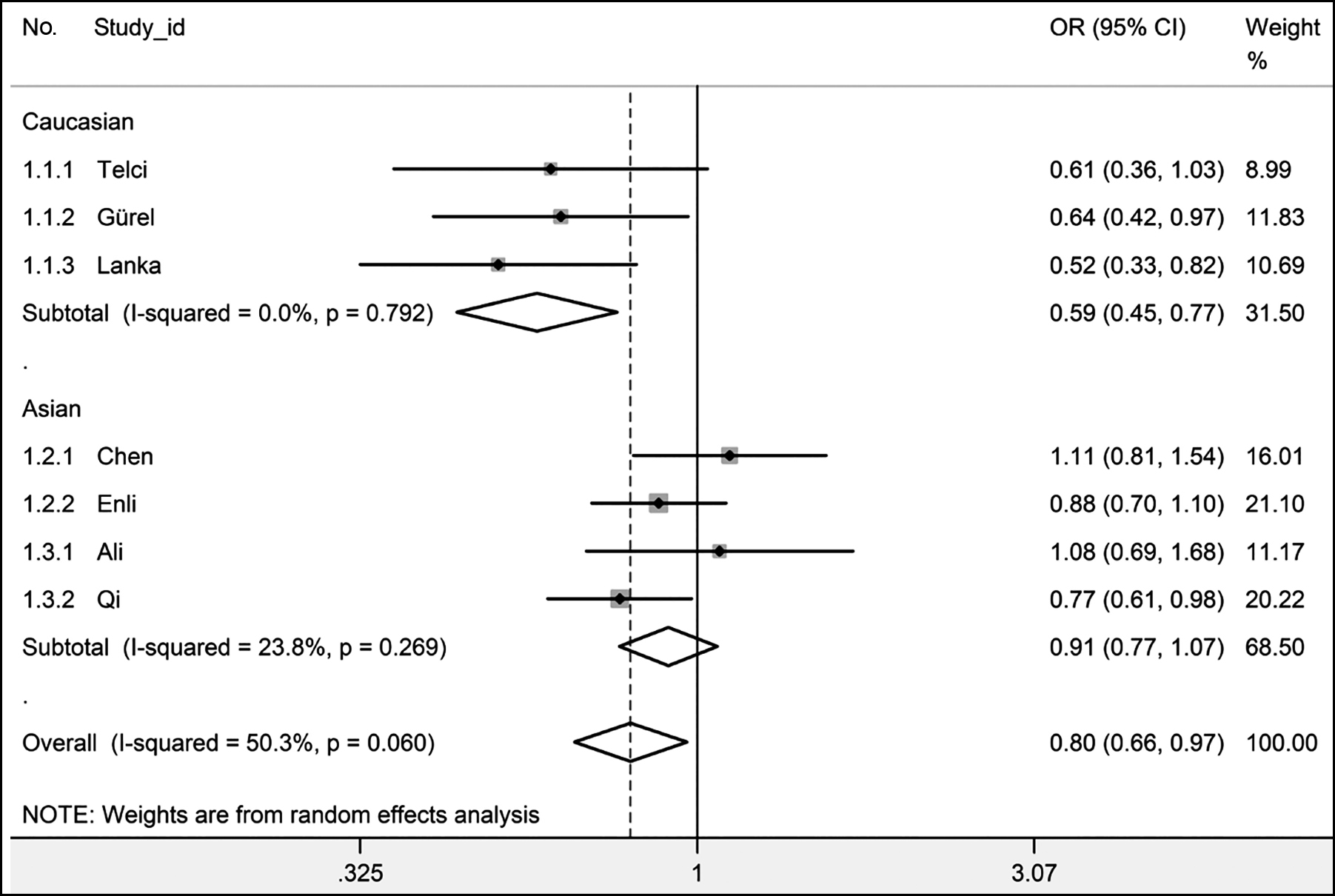 Figure 2: In allelic comparison (A vs. G) overall, forest plot for the association between Klotho rs1207568 and urolithiasis with a random-effects model. A box and a horizontal line means the estimate of the OR and its 95% CI for each study. Rhombus means pooled OR and 95% CI.
Figure 2: In allelic comparison (A vs. G) overall, forest plot for the association between Klotho rs1207568 and urolithiasis with a random-effects model. A box and a horizontal line means the estimate of the OR and its 95% CI for each study. Rhombus means pooled OR and 95% CI.
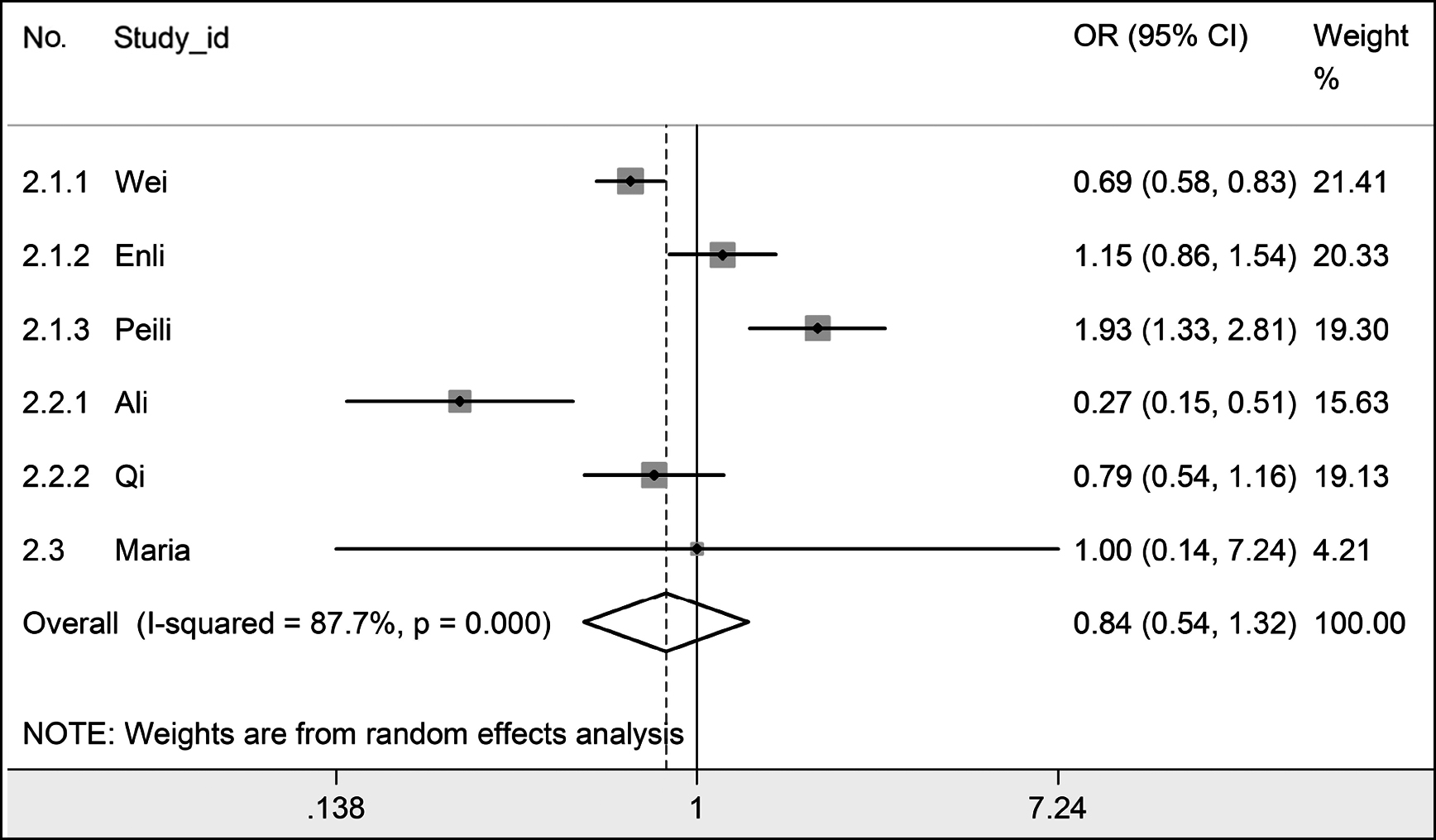 Figure 3: In allelic comparison (A vs. G) overall, the forest plot for the association between Klotho rs3752472 and urolithiasis with a random-effects model.
Figure 3: In allelic comparison (A vs. G) overall, the forest plot for the association between Klotho rs3752472 and urolithiasis with a random-effects model.
To evaluate the publication bias, Begg’s funnel plot and Egger’s test were used in any subgroup and any comparison of more than five studies. P-value of Begg’s test (PB), P-value of Egger’s test (PE), and symmetry of funnel plot were tested.13,14 According to the PB and PE value, no significant publication bias was discovered in each genetic models of rs1207568 overall, in each genetic models of rs3752472’s Asian subgroup, in allelic comparison (A vs. G) of rs3752472 overall, and in dominant model (AG+AA vs. GG) of rs564481 overall.
In the funnel plot, however, in the dominant model (AG + AA vs. GG) and heterozygote comparison (AG vs. GG) of rs1207568, study no 1.1.2, 1.1.3 and 1.3.1 extended beyond the diagonal line. The diagonal line indicated the pseudo-95% CI limit for the effect estimate. In allelic comparison (A vs. G), dominant model (AG + AA vs. GG), and heterozygote comparison (AG vs. GG) of rs3752472’s Asian subgroup, studies no 2.1.1, 2.1.3 and 2.2.1 extended beyond the diagonal line. In the homozygote comparison (AA vs. GG) of rs3752472’s Asian subgroup, study no 2.2.1 extended beyond the diagonal line. In allelic comparison (A vs. G) of rs3752472 overall, studies no 2.1.3 and 2.2.1 extended beyond the diagonal line.
DISCUSSION
Klotho rs1207568 adenine (A) was related to a decreased urolithiasis risk in the dominant model (AG + AA vs. GG), heterozygote comparison (AG vs. GG), and allelic comparison (A vs. G) in the Caucasian subgroup and overall; however, significant heterogeneity and unstable sensitivity analysis results were found for rs1207568 overall. In the Asian subgroup of rs1207568, unstable sensitivity analysis results were obtained by allelic comparisons (A vs. G). Publication bias and sensitivity analyses could not be made in the Han and Uyghur subgroups. The publication bias analysis results suggested differences between the subgroups. These results suggested that Klotho rs1207568 adenine (A) is related to a decreased urolithiasis risk in Caucasians. There were inadequate data to confirm the relation between urolithiasis susceptibility and Klotho rs1207568 in Asians, and the results should be interpreted with caution.
For Klotho rs3752472, heterogeneity was significant in each genetic model of the Han subgroup, and publication bias analyses could not be carried out in Han subgroup, but the results were stable in sensitivity analyses. Statistically significant changes were found in the recessive model (AA vs. GG + AG) and the homozygote comparison (AA vs. GG) of rs3752472 in the Uyghur subgroup; however, publication bias and sensitivity analyses could not be carried out. Only one study was included in the Caucasian subgroup of rs3752472. The publication bias analysis results suggested differences between the subgroups. These results showed that Klotho rs3752472 may not be related to the risk of urolithiasis in the Han subgroup of Asians. There were inadequate data to confirm the association between urolithiasis susceptibility and Klotho rs3752472 in Uyghur and Caucasians, and the results should be explained with caution.
No statistically significant change in urolithiasis risk was detected in any genetic model or subgroup for rs564481, rs650439, F352V, and rs145682430. In each study included in each SNP, no statistically significant changes were found. However, publication bias analysis could not be performed. Heterogeneity was not found in any genetic model or subgroup for rs564481, rs650439, or F352V. The results for rs564481 overall and the Asian subgroup of rs650439 showed stability in the sensitivity analyses. These results showed that Klotho rs564481 might not be related to urolithiasis risk in Asians or Caucasians, and that Klotho rs650439 might not be related to urolithiasis risk in Asians. There were inadequate data to confirm the relation between urolithiasis susceptibility and Klotho F352V in Caucasians or Klotho rs145682430 in Asians, and the results should be interpreted with caution.
Simultaneously, limitations of this meta-analysis should be addressed. To date, there had been few practical studies and their subgroups that could be absorbed by meta-analysis. In some groups or subgroups, sensitivity or publication bias analyses could not be operated. Studies no 1.1.1, 1.1.2, 1.1.3, 1.3.1 and 5.2 departed from HWE significantly. Unpublished studies or studies written by other languages were excluded. With imperfection, this meta-analysis and systematic review provided insights into the underlying relation between urolithiasis and Klotho gene polymorphisms.
CONCLUSION
Klotho rs1207568 adenine (A) may be related to a decreased urolithiasis risk in Caucasians. Klotho rs3752472 may not be related to urolithiasis risk in Han Asian subgroup. Klotho rs564481 may not be related to urolithiasis risk in Asians or Caucasians, and Klotho rs650439 may not be related to urolithiasis risk in Asians.
There were inadequate data to confirm the relation between urolithiasis susceptibility and Klotho rs1207568 in Asians, the relation between urolithiasis susceptibility and Klotho rs3752472 in Uyghur or Caucasians, and the relation between urolithiasis susceptibility and Klotho F352V in Caucasians or Klotho rs145682430 in Asians, and the results should be interpreted with caution. Elaborately designed studies with added subgroups and larger sample sizes will be needed to check the risk identified in systematic reviews and meta-analyses.
COMPETING INTEREST:
The authors declared that they have no competing interests.
AUTHORS’ CONTRIBUTION:
JQ, BD: Designed the study and drafted the manuscript, accumulated the data, analysis and interpretation of the data, substantively revised the manuscript.
JX: Designed the study and drafted the manuscript, subs-tantively revised the manuscript.
BC: Accumulated the data, conducted the analysis and inter-pretation of the data, substantively revised the manuscript.
TW: Accumulated the data, conducted the analysis and interpretation of the data.
All authors read and approved the final manuscript for publi-cation.
REFERENCES
- Danpure CJ. Genetic disorders and urolithiasis. Urol Clin North Am 2000; 27(2):287-99, viii. doi: 10.1016/s0094- 0143(05) 70258-5.
- Devuyst O, Pirson Y. Genetics of hypercalciuric stone forming diseases. Kidney Int 2007; 72(9):1065-72. doi: 10.1038/ sj.ki.5002441.
- Zhao Y, Zeng X, Xu X, Wang W, Xu L, Wu Y, et al. Low-dose 5-aza-2'-deoxycytidine protects against early renal injury by increasing Klotho expression. Epigenomics 2022; 14(22): 1411-25. doi: 10.2217/epi-2022-0430.
- Cuarental L, Ribagorda M, Ceballos MI, Pintor-Chocano A, Carriazo SM, Dopazo A, et al. The transcription factor Fosl1 preserves Klotho expression and protects from acute kidney injury. Kidney Int 2023; 103(4):686-701. doi: 10.1016/j.kint. 2022.11.023.
- Taguchi K, Yasui T, Milliner DS, Hoppe B, Chi T. Genetic risk factors for idiopathic urolithiasis: a systematic review of the literature and causal network analysis. Eur Urol Focus 2017; 3(1):72-81. doi: 10.1016/j.euf.2017.04.010.
- Erben RG, Andrukhova O. FGF23-Klotho signaling axis in the kidney. Bone 2017; 100:62-8. doi: 10.1016/j.bone.2016.09. 010.
- Kawano K, Ogata N, Chiano M, Molloy H, Kleyn P, Spector TD, et al. Klotho gene polymorphisms associated with bone density of aged postmenopausal women. J Bone Miner Res 2002; 17(10):1744-51. doi: 10.1359/jbmr.2002.17.10.1744.
- Xu C, Zhang W, Lu P, Chen JC, Zhou YQ, Shen G, et al. Mutation of Klotho rs3752472 protect the kidney from the renal epithelial cell injury caused by CaOx crystals through the Wnt/β-catenin signaling pathway. Urolithiasis 2021; 49(6):543-50. doi: 10.1007/s00240-021-01269-z.
- Qin J, Xing J, Liu R, Chen B, Chen Y, Zhuang X. Association between CD40 rs1883832 and immune-related diseases susceptibility: A meta-analysis. Oncotarget 2017; 8 (60):102235-43. doi: 10.18632/oncotarget.18704.
- Qin J, Cai Z, Xing J, Duan B, Bai P. Association between calcitonin receptor gene polymorphisms and calcium stone urolithiasis: A meta-analysis. Int Braz J Urol 2019; 45(5): 901-9. doi: 10.1590/S1677-5538.IBJU.2019.0061.
- GA Wells, B Shea, D O'Connell, J Peterson, V Welch, M Losos, et al. The Newcastle-Ottawa Scale (NOS) for assessing the quality of nonrandomised studies in meta-analyses. Available from: http://www.ohri.ca/programs/clinical_epide miology/nosgen.pdf (Accessed on 06/21/2017).
- Der Simonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials 1986; 7:177–88. doi: 10.1016/0197- 2456(86)90046-2.
- Begg CB, Mazumdar M. Operating characteristics of a rank correlation test for publication bias. Biometrics 1994; 50(4):1088-101.
- Egger M, Smith GD, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ 1997; 315(7109):629-34. doi: 10.1136/bmj.315.7109.629.
- Apolikhin OI, Sivkov AV, Konstantinova OV, Slominskij PA, Tupicyna TV, Kalinichenko DN. Genetic risk factors for multiple kidney stone formation in the russian population. Urologiia 2015; (4):4-6.
- Apolikhin OI, Sivkov AV, Konstantinova OV, Slominsky PA, Tupitsyna TV, Kalinichenko DN. Genetic risk factors for recurrence-free urolithiasis in the russian population. Urologiia 2016; (4):20-3.
- Apolikhin OI, Sivkov AV, Konstantinova OV, Slominskii PA, Tupitsyna TV, Kalinichenko DN. Early diagnosis of risk for developing calcium oxalate urolithiasis. Urologiia 2017; (3):5-8. doi: 10.18565/urol.2017.3.5-8.
- Telci D, Dogan AU, Ozbek E, Polat EC, Simsek A, Cakir SS, et al. Klotho gene polymorphism of G395A is associated with kidney stones. Am J Nephrol 2011; 33(4):337-43. doi: 10.1159/000325505.
- Gurel A, Ure I, Temel HE, Cilingir O, Uslu S, Celayir MF, et al. The impact of Klotho gene polymorphisms on urinary tract stone disease. World J Urol 2016; 34(7):1045-50. doi: 10.1007/s00345-015-1732-z.
- Lanka P, Devana SK, Singh SK, Sapehia D, Kaur J. Klotho gene polymorphism in renal stone formers from North western India. Urolithiasis 2021; 49(3):195-9. doi: 10.1007/ s00240-020-01226-2.
- Chen X, Rijin S, Xiaolan W, Wei Z. The association among vitamin D receptor gene, Klotho gene polymorphisms and calcium urolithiasis. Chin J Exp Surg 2013; 30(12):2554-7. doi: 10.3760/cma.j.issn.1001-9030.2013.12.024.
- Enli L. Study on Urinary stones genetic polymorphism. Tianjin Medical University; 2018.
- Ali A, Tursun H, Talat A, Abla A, Muhtar E, Zhang T, et al. Association Study of Klotho Gene polymorphism with calcium oxalate stones in the uyghur population of Xinjiang, China. Urol J 2017; 14(1):2939-43. doi: 10.22037/uj.v14i1. 3636.
- Qi S. Study on association of Klotho gene polymorphism and idiopathic calcium oxalate kidney stones in Uighur population of Xinjiang region. Xinjiang Medical University 2019. doi: 10.27433/d.cnki.gxyku.2019.000088.
- Wei X, Minjun J, Chen X, Rijin S, Xiaolan W, Wei Z. Klotho gene polymorphism of rs3752472 is associated with the risk of calcium oxalate calculi. J of Modern Urol 2015; 20(2):123-7. doi: 10.3969/j.issn.1009-8291.2015-02-016.
- Peili M, Xin Y, Jingjin G, Reheman A, Yi H, Ruotian L, et al. The relationship between Klotho gene and idiopathic calcium oxalate renal calculi in Uygur and Han nationalities in Xinjiang. J Modern Urol 2020; 25(9):778-83. doi: 10.3969/j.issn.1009-8291.2020.09.004.
- Litvinova MM, Khafizov K, Korchagin VI, Speranskaya AS, Asanov AY, Matsvay AD, et al. Association of CASR, CALCR, and ORAI1 genes polymorphisms with the calcium urolithiasis development in russian population. Front Genet 2021; 12:621049. doi: 10.3389/fgene.2021.621049.
- Liuya Y, Talat A, Guangjian D, Mahmut M. Study on association of calcium oxalate stones and Klotho gene polymorphism in Uighur population of Xinjiang region. J Clin Exp Med 2015; 14(3):183-5. doi: 10.3969/j.issn.1671- 4695.2015.03.005.