SHANK2-FGFR1: A Novel FGFR1 Rearrangement in a Patient with Intestinal Adenocarcinoma
By Hongguang Sheng1, Yugang Jiang1, Ling Li2Affiliations
doi: 10.29271/jcpsp.2021.11.1384
Sir,
The FGFR gene rearrangements, such as FGFR1, FGFR2 and FGFR3, have been found in a variety of cancers, including cholangiocarcinoma, bladder cancer, breast cancer, thyroid cancer, head and neck cancer, lung cancer, and liver cancer.1,2 FGFR1/2/3 fusions were detected in 3.5% of patients with squamous cell lung cancer and 1.3% of patients with non-small cell lung cancers (NSCLC).3 Here, we describe a novel FGFR1 rearrangement (SHANK2-FGFR1) in a intestinal adenocarcinoma patient.
A 55-year male presented to hospital with fever for more than two months. Colonoscopy showed multiple polyps of colon, and a polyp 20 cm away from anus was consistent with the change of intramucosal adenocarcinoma. Laparoscopic-assisted Dixon surgery was performed. A large tumor of 3×2 cm in the upper rectum was found during the surgery. The postoperative pathological examination showed protuberant type of well to moderately differentiated adenocarcinoma (Figure 1), and about 10% of the area was poorly differentiated adenocarcinoma; and the tumor invading the superficial muscle layer.
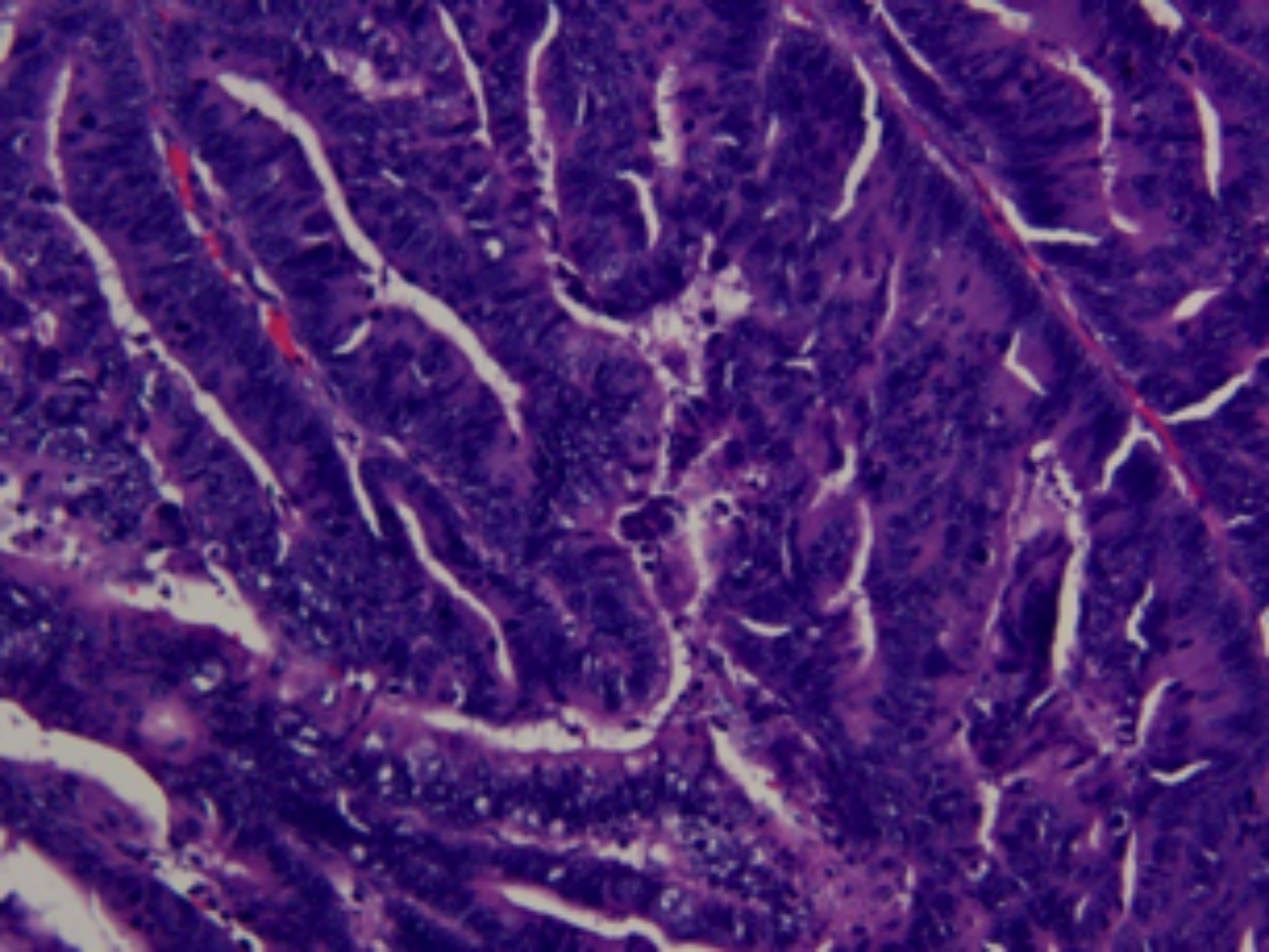 Figure 1: Histopathologic image of formalin fixed and paraffin embedded section of the tumor. The tumor is well to moderately differentiated adenocarcinoma. (H&E, ×200).
Figure 1: Histopathologic image of formalin fixed and paraffin embedded section of the tumor. The tumor is well to moderately differentiated adenocarcinoma. (H&E, ×200).
In order to find an accurate treatment, next-generation sequencing (NGS) analysis was performed on the paraffin-embedded specimens. The analysis revealed a novel SHANK2-FGFR1 fusion (Figure 2A). This SHANK2-FGFR1 fusion was generated by the fusion of exons 1–11 of SHANK2 to exons 9–24 of FGFR1 (Figure 2B). No other variants were found except for a concomitant FGFR1 and CCND1 amplification. SHANK2-FGFR1 fusion has not been reported so far in intestinal adenocarcinoma. This fusion retains the complete kinase domain of FGFR1 gene, which may lead to the activation of FGFR1 gene.
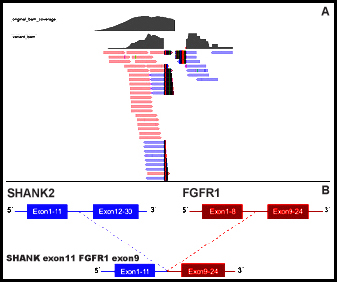 Figure 2: Next-generation sequencing results of SHANK2-FGFR1 fusion. (A) The snapshot of SHANK2-FGFR1 by Integrative Genomics Viewer. (B) Schematic representation of the domain structure of the SHANK2-FGFR1 fusion protein.
Figure 2: Next-generation sequencing results of SHANK2-FGFR1 fusion. (A) The snapshot of SHANK2-FGFR1 by Integrative Genomics Viewer. (B) Schematic representation of the domain structure of the SHANK2-FGFR1 fusion protein.
Tyrosine kinase inhibitors (TKIs) have been used successfully in chronic myeloid leukaemia (CML) treatment. Currently, several TKIs have been described as treatment alternatives in the 8p11 myeloproliferative syndrome (EMS).4,5 Among them, ponatinib is the third-generation TKI that can effectively inhibit the activity of kinases, such as Bcr-Abl/PDGFRA/FGFR/KIT. FDA has approved ponatinib for the treatment of FGFR alteration-positive CML and Philadelphia chromosome positive acute lymphoblastic leukemia (ALL) patients. Meanwhile, a clinical trial of ponatinib for the treatment of solid tumor patients with FGFR gene mutation/amplification/fusion is underway (NCT0 2272998). In addition, several novel FGFR inhibitors that selectively target FGFR1/2/3 have potential anti-tumor activity; and are currently in clinical trials. For example, the phase II clinical trial of BGJ398 for solid tumors carrying FGFR genomic variation has been completed (NCT02160041), and the NCI-MATCH clinical trial of AZD4547 is also being carried out (NCT024 65060).
In summary, this is the first report of a novel SHANK2-FGFR1 rearrangement in a patient with intestinal adenocarcinoma. Hopefully, this new type of fusion can be helpful for the future application of FGFR1 inhibitors to alleviate the progression of tumors after the patients fail in multi-line therapy.
ACKNOWLEDGMENT:
This work of partial data and collection was supported by Clinical Oncology Research Alliance (CORA).
CONFLICT OF INTEREST:
The authors declared no conflict of interest.
AUTHORS' CONTRIBUTION:
LL: Data analysis and manuscript writing.
HS: Research design and data analysis.
YJ: Edited and revised the manuscript.
REFERENCES
- Singh D, Chan JM, Zoppoli P, Niola F, Sullivan R, Castano A, et al. Transforming fusions of FGFR and TACC genes in human glioblastoma. Science 2012; 337(6099):1231-5. doi: 10.1126/science.1220834.
- Wu YM, Su F, Kalyana-Sundaram S, Khazanov N, Ateeq B, Cao X, et al. Identification of targetable FGFR gene fusions in diverse cancers. Cancer Discov 2013; 3(6):636-47. doi: 10.1158/2159-8290.CD-13-0050.
- Wang R, Wang L, Li Y, Hu H, Shen L, Shen X, et al. FGFR1/3 Tyrosine Kinase Fusions Define a Unique Molecular Subtype of Non–Small Cell Lung Cancer. Clin Cancer Res 2014; 20(15):4107-14. doi: 10.1158/1078-0432.CCR-14-0284.
- Abruzzese E, Breccia M, Latagliata R. Second-Generation Tyrosine Kinase Inhibitors in First-Line Treatment of Chronic Myeloid Leukaemia (CML). Bio Drugs 2014; 28(325):17-26. doi: 10.1007/s40259-014-0093-2.
- Jabbour E, Kantarjian H, Cortes J. Use of second- and third-generation tyrosine kinase inhibitors in the treatment of chronic myeloid leukemia: an evolving treatment paradigm. Clin Lymphoma Myeloma Leuk 2015; 15(6):323-34. doi: 10.1016/j.clml.2015.03.006.