Ki-67 Quantification in Breast Cancer by Digital Imaging AI Software and its Concordance with Manual Method
By Talat Zehra1, Mahin Shams2, Zubair Ahmad3, Qurratulain Chundriger3, Arsalan Ahmed3, Nazish Jaffar1Affiliations
doi: 10.29271/jcpsp.2023.05.544ABSTRACT
Objective: To validate the concordance of automated detection of Ki67 in digital images of breast cancer with the manual eyeball / hotspot method.
Study Design: Descriptive study.
Place and Duration of the Study: Jinnah Sindh Medical University, Karachi, from 1st January to 15th February 2022.
Methodology: Glass slides of cases diagnosed as invasive ductal carcinoma (IDC) were obtained from the Agha Khan Medical University Hospital, selected retrospectively and randomly from 60 patients. They were stained with the Ki67 antibody. An expert pathologist evaluated the Ki67 index in the hotspot fields using eyeball method. Digital images were taken from the hotspots using a camera attached to the microscope. The images were uploaded in the Mindpeak software to detect the exact percentage of Ki67-positive cells. The results obtained through automated detection were compared with the results reported by expert pathologists to see the differential outcome.
Results: The manual and automated scoring methods showed strong positive concordance (p <0.001).
Conclusion: Automated scoring of Ki-67 staining has tremendous potential as the issues of lack of consistency, reproducibility, and accuracy can be eliminated. In the era of personalised medicine, pathologists can efficiently give a precise clinical diagnosis with the support of AI.
Key Words: Artificial intelligence, Algorithms, Breast cancer, Deep learning, Image detection, Ki-67.
INTRODUCTION
Invasive ductal carcinoma (IDC) of breast is the most common malignant tumour in women worldwide. According to GLOBOCAN 2020, female breast cancer has become the most commonly diagnosed cancer replacing lung cancer. Annual new cases of breast cancer are estimated to be 2.3 million (11.7%) followed by lung cancer (11.4%). The mortality rate for breast cancer is estimated to be 6.9%.1,2
The main biological indicators for molecular typing, treatment and evaluation of the prognosis of breast carcinoma are Ki67, Estrogen Receptor (ER), Progesterone receptor (PR), and Her2neu. Ki67 is a marker that is used as an indicator of cell proliferation in cancer. It is expressed at G1, S, G2, and M phases of tumour cells. Ki67 scoring is important in the grading and determination of prognosis and treatment of invasive ductal carcinoma of breast.3-5
Ki67 assessment remains a challenge due to intra and inter-observer variability and bias in the manual hotspot scoring method.5 This has raised concerns regarding the validity of this marker as a prognostic indicator and the impact it can have on decision-making regarding therapy for breast cancer.5,6 Using automated method for Ki67 scoring, problems related to lack of consistency, reproducibility, and selection bias can be addressed.7,8 Human errors can be minimised thus helping the pathologists to deliver accurate reports using tissue samples. However, robust detection and segmentation of cancer cell nuclei are important for the assessment of Ki67 by automated methods.9,10
Ki67 staining and scoring assessment is closely related to the clinical, pathological, and molecular features of breast cancer and can be used to guide the prognosis and treatment of breast cancer patients.10-12 Ki-67 scoring can also be used to distinguish luminal breast cancer subtypes and determine personalised treatment for each particular case.10-14
Whole slide digital scanning and histopathological image analysis combined with artificial intelligence (AI) technology can provide high definition and high-speed analysis thus aiding the pathologists in providing accurate results.15,16 Extensive literature search was performed to find a local study performed to compare Ki 67 quantification by manual versus automation methods, failing which, this study was designed with the objective to compare the automated detection of Ki67 with the manual eyeball / hotspot method.
METHODOLOGY
Slides of cases previously diagnosed as IDC of breast were obtained from the Section of Histopathology, Department of Pathology and Laboratory Medicine, The Aga Khan University Hospital, after taking ethical approval from the hospital’s Ethical Review Committee. The slides were selected retrospectively and randomly from 60 patients previously diagnosed as cases of breast cancer during the period of one and a half months from 1st January to 15th February 2022.
The slides were stained with the Ki 67 antibody. An expert pathologist independently evaluated the Ki67 index in the hotspot fields using the manual eyeball method. Digital images were taken from the hotspot areas of tumour using a camera attached to the microscope. All slides were digitalised at 40x (Figure 1). The images were uploaded in the Mindpeak software to detect Ki67-positive tumour cells as red dots. Ki67-negative tumour cells were detected as yellow dots. Also, software was able to give the exact percentage of Ki 67 positive cells in a selected region (Figures 2 and 3). The results obtained through automated detection were compared with the results given by expert pathologists to see the differential outcome. Slides having poor fixation or poorly identified areas of IDC were excluded. Poorly stained slides were also excluded.
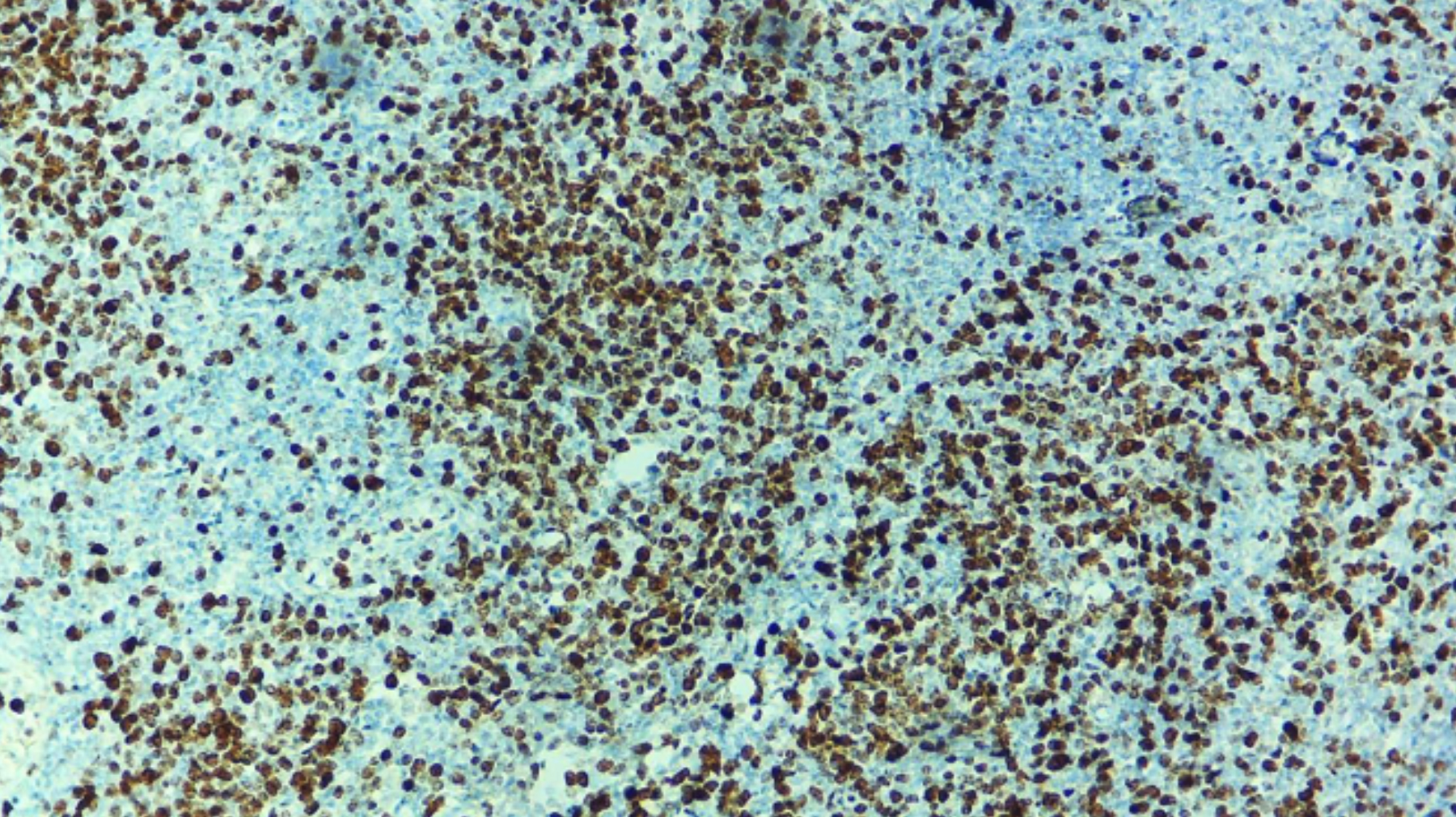 Figure 1: The digital image made at 40x by microscope connected camera. Manual scoring was performed before uploading it to the software. Manual quantification was performed independently by a consultant histopathologist.
Figure 1: The digital image made at 40x by microscope connected camera. Manual scoring was performed before uploading it to the software. Manual quantification was performed independently by a consultant histopathologist.
The results obtained through automated detection were compared with the results reported by expert pathologists to see the concordance. Data were analysed using Statistical Package for Social Sciences (SPSS) 20.0. A paired t-test was applied to see the concordance between the two methods. A p-value of <0.001 was considered significant.
RESULTS
Mean values calculated for Ki67 quantification by both manual and automated AI-based methods were 36.40±25.7 and 38.35±24.7, respectively. The manual and automated scoring methods showed strong concordance as the paired sample t-test was significant (0.00). The p-value was <0.001 indicating a statistical significance. The software was not only able to quantify tumour-positive cells but also tumour-negative cells and also give the exact percentage of tumour-positive cells in selected fields (Figures 1-3).
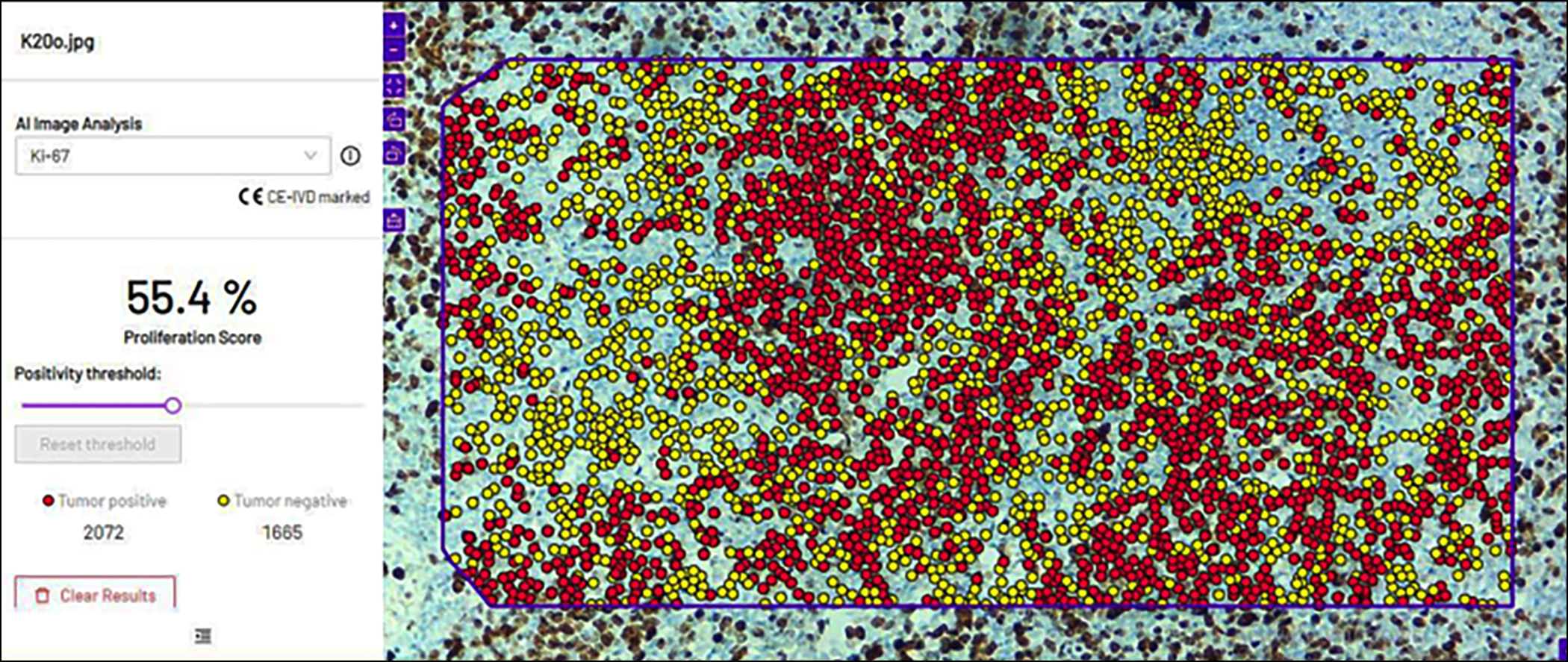 Figure 2: After uploading the figure 1 image in the software, it showed red dots for positive tumour cells for Ki-67 and yellow dots for negative tumour cells. On the right, the software gave the quantification and percentage of both tumour-positive and tumour-negative cells in a selected region.
Figure 2: After uploading the figure 1 image in the software, it showed red dots for positive tumour cells for Ki-67 and yellow dots for negative tumour cells. On the right, the software gave the quantification and percentage of both tumour-positive and tumour-negative cells in a selected region.
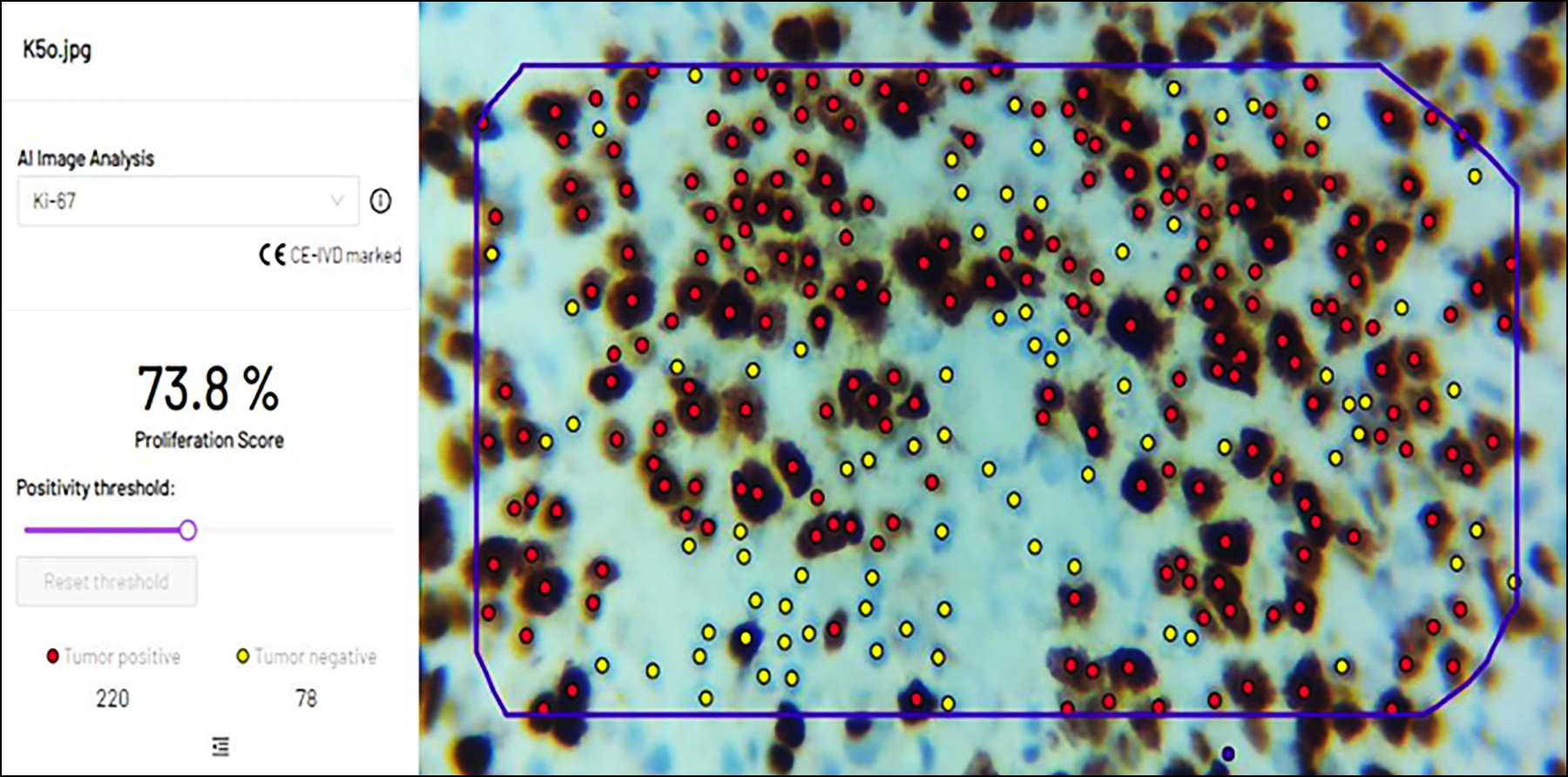 Figure 3: In another image taken by camera-connected microscope, software was able to identify all tumour positive and negative cells along with percentages in the selected region.
Figure 3: In another image taken by camera-connected microscope, software was able to identify all tumour positive and negative cells along with percentages in the selected region.
DISCUSSION
Breast cancer is a complex disease with various subtypes, and treatment decisions often rely on the accurate pathological evaluation of the tumour. One of the key factors used in breast cancer treatment and the prognosis is the Ki67 proliferation index, which reflects the percentage of tumour cells that are actively dividing.2,14 While manual assessment of Ki67 has been the gold standard for several years, automated scoring methods using digital pathology and AI are gaining traction due to their potential for higher reproducibility, and efficiency and for strengthening the role of pathologists in computer-aided diagnosis.17-19
According to the International Ki-67 in Breast Cancer Working Group guidelines for the analysis, reporting and use of Ki-67, the Ki-67 score is defined as the percentage of invasive cancer cells stained positively in the examined region. The intensity of staining is not relevant in Ki 67 staining. Both core biopsies and whole section tissues are suitable. Current guidelines recommend that at least three high power fields (HPFs) should be selected which should be representative of the staining in the entire section.3,20-22 One important reason for the poor reproducibility of Ki67 scoring is the inconsistency in selecting the representative area on the slide. The cancer cells are not evenly distributed on the slide. There are hotspot areas where most ki67-positive tumour cells are concentrated and there are other areas with few Ki67 positive tumours cells.2,13 A number of studies have shown that the Ki-67 score assessed in only the hotspot area or margin area is significantly higher than the average area. Also, the Ki-67 score in the hotspot area has a greater correlation with the prognosis of breast cancer.9,10,11 The number of cells counted on the same section also affects the reproducibility of Ki-67 scoring in breast cancer. The International Ki-67 Working Group has recommended that at least 1000 cells should be scored.2,9,23
Several studies have compared manual and automated Ki67 scoring in breast cancer, with varying results. A study in 2021 found that automated scoring had an excellent correlation with manual scoring and was faster and more objective, suggesting that automated scoring could improve diagnostic reproducibility, potentially reduce interobserver variability, and facilitate clinical decision-making.19
However, on the contrary, other studies have reported lower accuracy with automated Ki67 scoring. A study found that while automated scoring had high agreement with manual scoring in certain cases, it was less accurate in cases with high heterogeneity or when the Ki67 staining was not uniform.23
However, it is agreed upon by various studies that straightforward guidelines need to be developed for Ki67 biomarker quantification in breast tumour tissues by taking into consideration the pre-analytical as well as analytical phases of the laboratory procedures. Furthermore, it is an established fact now that AI-based digital methods can help mitigate the pathologists’ workload in dealing with repetitive complex tasks and their precious time can be utilised for complex cases and important decision-making. Experimental AI-based studies require validation for their optimal utilisation in clinical practice.5,6,19
The use of artificial intelligence in histopathology is becoming well-established with every passing day. AI techniques will not replace pathologists at all but will infact give pathologists an even more important and commanding position.24,25 By using these techniques the role of a pathologist will not just be supportive, he or she will be in a position to make disease models, determine disease trends, and predict disease outcomes according to demographic region, hence, heralding a new era of personalised medicine.25 With an alarming increase in cancer globally which is putting a tremendous load on pathologists whose number is declining, these innovative modalities can make a significant difference and result in marked improvement in patient outcomes.
This study was limited by the absence of whole slide scanner or digital microscope, as only digital images were created rather than whole slide images. The algorithm can only be used in breast cancer cases and not in other tumours of the body. Another limitation was that the current software worked best with 40X magnification.
In the era of personalised medicine, pathologists can efficiently give a precise clinical diagnosis with the support of AI. Large validation studies on different organ-specific cohorts are required before clinical implementation. Multi-organ spectrum algorithms need to be developed.
CONCLUSION
The current study shows a high concordance with a significant p-value between the manual and automated scoring methods. Automated AI-based scoring of Ki-67 staining is substantial and has tremendous potential in the future increasing its clinical value as problems of consistency, reproducibility, and accuracy can be eliminated with the use of this novel modality.
ACKNOWLEDGEMENT:
We are thankful to Mindpeak GmbH software who provided the demo version for conducting this research.
ETHICAL APPROVAL:
Ethical approval was obtained from Ethical Review Committee, The Aga Khan University (2022-7757-21995).
PATIENTS’ CONSENT:
Not applicable.
COMPETING INTEREST:
The authors declared no competing interest.
AUTHORS’ CONTRIBUTION:
TZ: Conceived the idea, made digital slides, uploaded them in software, and analysed them.
MS: Wrote the manuscript, manually scored the digital images and entered the data for analysis.
QC, AA: Collected the Ki-67 slides of previously diagnosed cases of breast cancer patients data.
ZA: Critically revised the manuscript and did final corrections.
NJ: Analysed the data and prepared the results.
All the authors have approved the final version of the manuscript to be published.
REFERENCES
- World Health Organization. GLOBOCAN 2020; All cancers fact sheet. [Internet; cited June 2021]. Available from: https://gco.iarc.fr/today/data/factsheets/cancers/39-All-cancers-fact-sheet.pdf
- Pilleron S, Sarfati D, Janssen-Heijnen M, Vignat J, Ferlay J, Bray F, et al. Global cancer incidence in older adults, 2012 and 2035: A population-based study. Int J Cancer 2019; 144(1):49-58. doi.org/10.1002/ijc.31664.
- Nielsen TO, Samuel C, Leung Y, Rimm DL, Dodson A, Acs B, et al. Assessment of Ki67 in breast cancer: Updated recommendations from the international Ki67 in breast cancer working group. J Nat Cancer Inst 2021; 113(7): 80819. doi.org/10.1093/jnci/djaa201.
- Reisenbichler ES, Lester SC, Richardson AL, Dillon DA, Ly A, Brock JE. Interobserver concordance in implementing the 2010 ASCO/CAP recommendations for reporting ER in breast carcinomas: A demonstration of the difficulties of consistently reporting low levels of ER expression by manual quantification. Am J Clin Pathol 2013; 140(4): 487-94. doi: 10.1309/AJCP1RF9FUIZRDPI.
- Gudlaugsson E, Skaland I, Janssen EA, Smaaland R, Shao Z, Malpica A, et al. Comparison of the effect of different techniques for measurement of Ki67 proliferation on reproducibility and prognosis prediction accuracy in breast cancer. Histopathol 2012; 61(6):1134-44. doi: 10.1111/j. 1365-2559.2012.04329.x.
- Robertson S, Azizpour H, Smith K, Hartman J. Digital image analysis in breast pathology-from image processing techniques to artificial intelligence. Transl Res 2018; 194: 19-35. doi: 10.1016/j.trsl.2017.10.010.
- Ibrahim A, Gamble P, Jaroensri R, Abdelsamea MM, Mermel CH, Chen PC, et al. Artificial intelligence in digital breast pathology: Techniques and applications. Breast 2020; 49:267-73. doi: 10.1016/j.breast.2019.12.007.
- Barisoni L, Hodgin JB. Digital pathology in nephrology clinical trials, research, and pathology practice. Curr Opin Nephrol Hypertens 2017; 26(6):450-9. doi: 10.1097/MNH. 0000000000000360.
- Arima N, Nishimura R, Osako T, Nishiyama Y, Fujisue M, Okumura Y, et al. The importance of tissue handling of surgically removed breast cancer for an accurate assessment of the KI-67 index. J Clin Pathol 2016; 69(3):255-9. doi: 10.1136/jclinpath-2015-203174.
- Yuan P, Xu B, Wang C, Zhang C, Sun M, Yuan L. Ki-67 expression in luminal type breast cancer and its association with the clinicopathology of the cancer. Oncol Lett 2016; 11(3):2101-5. doi: 10.3892/ol.2016.4199.
- Miller HC, Drymousis P, Flora R, Goldin R, Spalding D, Frilling A. Role of KI-67 proliferation index in the assessment of patients with neuroendocrine neoplasias regarding the stage of disease. World J Surg 2014; 38(6):1353-61. doi: 10.1007/s00268-014-2451-0.
- Rademakers SE, Hoogsteen IJ, Rijken PF, Terhaard CH, Doornaert PA, Langendijk JA, et al. Prognostic value of the proliferation marker KI-67 in laryngeal carcinoma: Results of the accelerated radiotherapy with carbogen breathing and nicotinamide phase III randomized trial. Head Neck 2015; 37(2):171-6. doi: 10.1002/hed.23569
- Mungle T, Tewary S, Arun I, Basak B, Agarwal S, Ahmed R, et al. Automated characterization and counting of KI-67 protein for breast cancer prognosis: A quantitative immunohistochemistry approach. Comput Methods Prog Biomed 2017; 139:149-61. doi: 10.1016/j.cmpb.2016.11. 002.
- Coates AS, Winer EP, Goldhirsch A, Gelber RD, Gnant M, Piccart-Gebhart MJ, et al. Tailoring therapies-improving the management of early breast cancer: St Gallen international expert consensus on the primary therapy of early breast cancer 2015. Ann Oncol 2015; 26(8):1533-46. doi: 10.1093/ annonc/mdv221.
- Wang W, Wu JY, Zhang PF, Fei XC, Zong Y, Chen XS, et al. Prognostic and predictive value of KI-67 in triple-negative breast cancer. Oncotarget 2016; 7(21):31079-87. doi: 10.18632/oncotarget.9075.
- Irshad H, Veillard A, Roux L, Racoceanu D. Methods for nuclei detection, segmentation, and classification in digital histopathology: A review-current status and future potential. IEEE Rev Biomed Eng 2014; 7:97-114. doi: 10. 1109/RBME.2013.2295804.
- Lloyd MC, Johnson JO, Kasprzak A, Bui MM. Image analysis of the tumour microenvironment. Adv Exp Med Biol 2016; 936:1-10. doi: 10.1007/978-3-319-42023-3_1.
- Uppu S, Krishna A. A deep hybrid model to detect multi-locus interacting SNPs in the presence of noise. Int J Med Inform 2018; 119:134-51. doi: 10.1016/j.ijmedinf.2018. 09.003.
- Boyaci C, Sun W, Robertson S, Acs B, Hartman J. Independent clinical validation of the automated Ki67 scoring guideline from the International Ki67 in Breast cancer working group. Biomolecules 2021 Oct 30; 11(11):1612. doi: 10.3390/biom11111612.
- Kermany DS, Goldbaum M, Cai W, Valentim CCS, Liang HY, Baxter SL, et al. Identifying medical diagnoses and treatable diseases by image-based deep learning. Cell 2018; 172(5):1122-31. doi: 10.1016/j.cell. 2018.02.010.
- Zhu C, Song F, Wang Y, Dong H, Guo Y, Liu J. Breast cancer histopathology image classification through assembling multiple compact CNNs. BMC Med Inform Decis Mak 2019; 19(1):198-214. doi: 10.1186/s12911-019-0913-x.
- Steiner DF, MacDonald R, Liu Y, Truszkowski P, Hipp JD, Gammage C, et al. Impact of deep learning assistance on the histopathologic review of lymph nodes for metastatic breast cancer. Am J Surg Pathol 2018; 42(12):1636-46. doi: 10.1097/PAS.0000000000001151.
- Hida AI, Omanovic D, Pedersen L, Oshiro Y, Ogura T, Nomura T, et al. Automated assessment of Ki-67 in breast cancer: the utility of digital image analysis using virtual triple staining and whole slide imaging. Histopathol 2020; 77(3):471-80. doi: 10.1111/his.14140.
- Zehra T, Shaikh A, Shams M. Dawn of artificial intelligence-enable digital pathology in Pakistan-A paradigm shift. J Pak Med Assoc 2021; 71(11):2683-4. doi: 10.47391/JPMA. 013257.
- Zehra T, Shabbir A. Adoption of digital pathology in developing countries: From benefits to challenges. J Coll Physicians Surg Pak 2021; 31(9):1120-2. doi: 10.29271/ jcpsp.2021.09.1120.