Increasing Frequency of New Delhi Metallo-beta-Lactamase and Klebsiella pneumoniae Carbapenemase Resistant Genes in a Set of Population of Karachi
By Faisal Iqbal Afridi1, Aliya Irshad Sani2, Rizma Khan3, Saeeda Baig2, Syed Aqib Ali Zaidi3, Qamar Jamal4Affiliations
doi: 10.29271/jcpsp.2023.01.63ABSTRACT
Objective: To determine the frequency of Klebsiella pneumoniae Carbapenemase (blaKPC) and New Delhi Metallo-Beta-Lactamase (blaNDM) resistant genes among clinical isolates of Enterobacterales in a set of Karachi population.
Study Design: An observational study.
Place and Duration of Study: Department of Microbiology, Dr. Ziauddin University Hospital, Karachi, Pakistan, from January 2019 to December 2020.
Methodology: A total of 2100 clinical isolates of Enterobacterales were collected. All isolates of Carbapenem-Resistant Enterobacterales (CRE) (Escherichia coli, Enterobacter and Klebsiella species) on the basis of Meropenem screening test positivity were included in the study. DNA was extracted and PCR was performed for resistant genes detection. Frequencies and percentages were computed for categorical variables and mean values and standard deviation for quantitative variables.
Results: Among 2100 isolates of Enterobacterales, the majority were E. coli 1260 (60%), followed by Klebsiella species 462 (22%), and Enterobacter species 210 (10%). The sources of CRE isolates included 34 (25%) from respiratory (tracheal aspirate, pleural fluid, and gastric lavage); 33 (24.26%) urine, 32 (25.53%) pus, 15 (11.03%) blood, and 20 (14.7%) others (ascitic fluid, stents, and tissue). All isolates of CRE were sensitive (100%) to Colistin, Tigecycline and Fosfomycin. Biochemically confirmed CRE 136 (6.5%) isolates, (79 (58%) males and 57 (42%) females), were selected for detecting resistant genes. The PCR showed 32 (23.52%) positive for both NDM and KPC resistant genes, 28 (20.58%) for NDM and 19 (13.97%) for KPC alone. Out of 79 followed up patients, 58 (73.4%) expired while 21 (26.6%) were discharged.
Conclusion: The frequency of blaNDM and blaKPC resistant genes in CRE isolates depicted increasing trend. Colistin, Fosfomycin, and Tigecycline showed high antimicrobial sensitivities in vitro. Further measures need to be applied for CRE with comprehensive resistant genes detection to curtail antimicrobial resistance.
Key Words: Frequency, KPC, NDM, Klebsiella species, Carbapenemases, Enterobacterales E.coli.
INTRODUCTION
Enterobacterales is a family of gram-negative organisms that cause a wide variety of infections of different parts in the body including urinary tract, respiratory tract, peritoneal cavity, and blood.1
Historically, these organisms have been readily treatable with antibiotics, but over the last several years resistance to extended-spectrum Cephalosporins (Cefotaxime or Ceftriaxone etc) has become a serious concern. Carbapenems have been reported to show effective antimicrobial activity and are considered as a preferred drug against ESBL-producing organisms such as Klebsiella pneumoniae (K. pneumoniae) and Escherichia coli (E. coli).2 Carbapenemases have three molecular classes which are A, B, and D. The class A is the most common and belongs to K. pneumoniae Carbapenemases (KPC). The class B contains New Delhi Metallo-beta-lactamases (NDM -1) enzymes which were first detected in New Delhi, India in antibiotic-resistant infections.3 Approximately thirty to fifty percent of Carbapenem-Resistant Enterobacterales (CRE) collected from UK, Pakistan, and India were NDM -1 producers.4 The overall mortality from invasive CRE can extend up to forty percent.5 However, there is a lack of updated statistical data within Pakistan and the exact frequency and prevalence of NDM-1 and KPC producers are not known.
Accurately identifying CRE in the clinical laboratory is an important first step in prevention. It is also important to understand how common CRE are with these resistant genes in the local healthcare setup. Through this study, one can implement aggressive infection control measures, antimicrobial stewardship, and increasing laboratory capacities to control the spread of these difficult-to-treat pathogens. This will also help to control the mortality rates due to infections with these superbugs. This study will be helpful in increasing awareness among medical professionals, the scientific community, and policymakers about the recent trend in antimicrobial resistance and the need for solutions by restricting the use of antimicrobials and switching to alternative options like screening for resistant genes as routine laboratory investigation. Data from this analysis can be used to guide how and where these strategies can be most efficiently implemented in Pakistan. The objective of the study was to determine the frequency of Klebsiella pneumoniae Carbapenemase (blaKPC) and New Delhi Metallo-Beta-Lactamase (blaNDM) resistant genes among clinical isolates of Enterobacterales in a set of Karachi population.
METHODOLOGY
This observational study was conducted from January 2019 to December 2020, in the Department of Clinical Microbiology of Ziauddin Medical University Hospital, Karachi. A total of 136 biochemically confirmed and Meropenem screen test positive isolates from Dr. Ziauddin University Hospital were included for the detection of blaKPC and blaNDM. Isolates from out-patients, repeated and duplicate isolates were excluded. Ethical approval of the study was obtained from the Ziauddin Ethics Review Committee. Informed consent was taken from either the patients or their close relatives. Three microbial species were identified i.e., E. coli, Enterobacter species and Klebsiella species in samples obtained from patients from ascitic fluid, bile, blood, CVP, empyema, gastric aspirate, peritoneal fluid, pus, sputum, stent, tissue, tracheal aspirate, and urine. The samples were received by the microbiology laboratory in sterile containers or in an Amies transport medium. In accordance with the standard microbiological techniques, these samples were processed and incubated at 35oC ± 2oC in ambient air for 24–28 hours for the growth of Enterobacterales. The members of Enterobacterales were recognised by using conventional techniques including colony morphology, gram staining, cytochrome oxidase test, differential growth on MacConkey’s agar medium and routine biochemical tests with additional usage of API 20E.6 Antimicrobial susceptibility testing was performed based on Clinical and Laboratory Standards Institute (CLSI) 2021 guidelines on Mueller Hinton agar (MHA) medium (Oxoid Ltd., England) using modified Kirby Bauer’s disk diffusion method.7 A 0.5 McFarland equivalent suspension of organism was prepared and inoculated onto MHA plates with subsequent application of antimicrobial discs. The plates were then incubated overnight at 35o5C ± 2oC in an ambient air incubator.
Carbapenem resistance was detected by screening test using Meropenem (10 µg) disk. Isolates showing Meropenem zone of inhibition of ≤19 mm were considered as resistant, while zone of inhibition of ≥23 mm was considered as sensitive, shown in Figure 1.
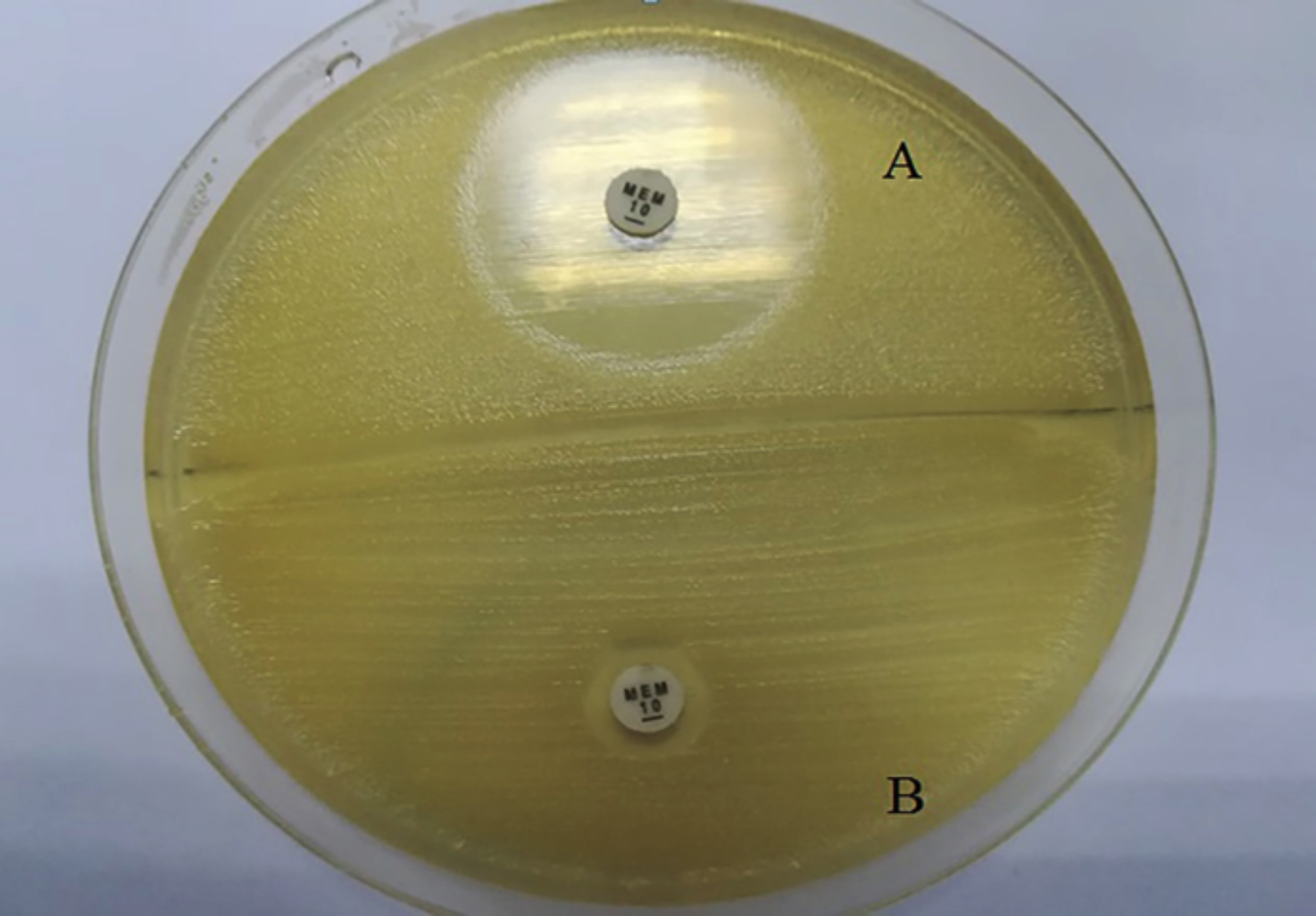 Figure 1: Meropenem disk screening test (A); Meropenem zone of inhibition of ≥ 23 mm. (B); Meropenem zone of inhibition of ≤ 19 mm.
Figure 1: Meropenem disk screening test (A); Meropenem zone of inhibition of ≥ 23 mm. (B); Meropenem zone of inhibition of ≤ 19 mm.
Antimicrobial susceptibility results were interpreted according to CLSI 2021 criteria. E. coli American Type Culture Collection (ATCC®) 25922, E. coli ATCC® 35218, and Pseudomonas aeruginosa ATCC® 27853 were used as control strains. The antimicrobial sensitivity testing of Colistin was performed by the determination of Minimum Inhibitory Concentration (MIC) by agar dilution method and interpreted according to EUCAST guidelines 2020 criteria. The MIC of <2 µg/ml was considered as sensitive, while the MIC of >2 µg/ml was considered as resistant. (https://www.eucast.org/ast_of_bacteria/previous_versions_ of_documents/). Tigecycline disk diffusion sensitivity criteria for Enterobacterales was taken as ≥19 mm (sensitive). (http:// www.accessdata.fda.gov/drugsatfda_docs/label/ 2009/021821 s017s018lbl.pdf).
From CRE-positive isolates, a single colony from pure culture was inoculated in TB (Tryptone Broth) for DNA extraction and incubated for 16 hours at 37oC. DNA extraction of isolates was carried out through spin column method, using a commercially available GeneJET Genomic DNA Purification Kit (cat: K0721, Thermo Fisher Scientific, USA). The procedure was followed according to manufacturer-provided protocol. All the extracted DNA samples were then stored at -20oC for further processing. The quality of extracted DNA was checked by a gold standard method of agarose gel electrophoresis.
Eluted DNA was quantified by Qubit 4 Fluorometer (Invitrogen by Thermofisher Scientific, USA). Qubit 4 Fluorometer was first calibrated by standard 1 and standard 2 provided by the Qubit 1X dsDNA assay (Invitrogen by Thermofisher Scientific, USA), 10 ul of each standard was added in respective labelled tube containing 190 ul of working buffer and both the tubes then streamed to read process. Once the Qubit 4 Fluorometer was calibrated, samples were formulated by adding 2 ul of sample and 198 ul of working buffer (dilution 1:100) followed by tube reading and quantification on Qubit 4 Fluorometer.
Genetic sequences of blaKPC and blaNDM were retrieved from the National Center for Biotechnology Information (NCBI). blaNDM gene sequence was downloaded from; https://www.ncbi. nlm.nih.gov/nuccore/NG_049326.1?report=fasta. The sequence for blaKPC was downloaded from https://www.ncbi.nlm.nih.gov/ nuccore/NG_049253.1?report=fasta. The sequence-specific primers for targeted region of blaKPC and blaNDM were designed by using an online available software Primer 3 (https://primer3. ut.ee/). Primers were commercially synthesised by acquiring services of Eurofins genomics (Germany). Details of primers is as follows:
blaNDM Forward 5’-GAAGCTGAGCACCGCATTAG-3’
Reverse 5’-GGGCCGTATGAGTGATTGC-3’
blaKPC Forward 5’-ATCGCCGTCTAGTTCTGCTG-3’
Reverse -5’-CGCTGTGCTTGTCATCCTT-3’
The detection of blaKPC and blaNDM was carried out by a acquiring a conventional PCR strategy.10 For this purpose, a commercially available DreamTaq Green PCR Master Mix 2X (K1081, Thermo ScientificTM) was used with different concentrations of primer and sample, to achieve an optimized protocol for both genes separately in CFX96TM (Bio-Rad, USA) thermocycler by using the following program: initial denaturation step 95oC for 5 min, followed by 35 cycles with denaturation at 95oC for 30 sec; annealing for 35 secs; at 59oC for blaNDM and 60oC for blaKPC; extension, 72oC for 35 sec and subsequently final extension 72oC for 7 minutes.
Amplified products of both genes were run on 1.5% agarose gel. The gel was visualised on Gel DocTM EZ Imager (Bio-Rad, USA) and image was analysed by ImageLab Software (Bio-Rad, USA). Data analysis was performed by using Statistical Package for Social Sciences (SPSS) version-25. Frequencies and percentages were computed for the presentation of all categorical variables like microorganisms, gender, comorbidities, antibiotics sensitivity, and resistance. Mean values and standard deviation were calculated for quantitative variables like the age of patients.
All the samples were obtained via clinical practice procedures and submitted in laboratory. Before analysis, the results were anonymised and no socio-demographic variables are analysed.
RESULTS
A total of 2100 clinical isolates of Enterobacterales were attained during the study period. Among these 2100 isolates, the majority of isolates were identified as E.coli 1260/2100 (60%), followed by Klebsiella species 462/2100 (22%), Enterobacter species 210/2100 (10%), Proteus species 63/2100 (3%), Morganella morgannii 21/2100 (1%), and others 84/2100 (4%). Predominantly, the isolates were from female patients 1300/2100 (61.9%), while the isolates from male patients were 800/2100 (38.1%). Female-to-male ratio was 1.6:1. The overall frequency of CRE among isolates of Enterobacterales was 136/2100 (6.5%). Out of 136 CRE- positive isolates, 79 (58%) were males and 57 (42%) were females. The frequency of CRE- positive among individual organism groups in CRE positive isolates was highest among E. coli n=52 (38.24 percent), Klebsiella species n=43 (31.62 percent), and Enterobacter species n=41 (30.15 percent).
According to the sources, of the 136 screened CRE isolates, 34 (25%) were from respiratory samples such as tracheal aspiration, pleural fluid and gastric lavage; 33 (24.26%) were obtained from urine, 32 (25.53%) from pus, 15 (11.03%) from blood, and 20 (14.7%) from other clinical samples. The other clinical samples included were ascitic fluid, stents, and tissue.
Antimicrobial susceptibilities results depicted that Aztreonam, Cefoperazone/sulbactam, Amoxicillin, Co-amoxiclav, and Meropenem were all resistant (0%) in CRE. On the other hand, all isolates of CRE were sensitive (100%) to Colistin, Tigecycline, and Fosfomycin. The resistance pattern of other antibiotics in clinical isolates of CRE is shown in Table I. Ciprofloxacin and Co-trimoxazole demonstrated resistance in more than 90% in all three organisms with overall 98% and 96%, respectively. However, Amikacin and Chloramphenicol showed comparatively lower resistance, 63% and 59%, respectively.
The PCR was performed on 136 isolates to detect NDM and KPC genes, out of which 57 (41.91%) samples were negative of either NDM or KPC. On the other hand, 32 (23.52%) showed presence of both resistant genes i.e., NDM and KPC. Twenty-eight (20.58%) samples were positive for NDM alone while 19 (13.97%) samples showed presence of KPC resistant gene.
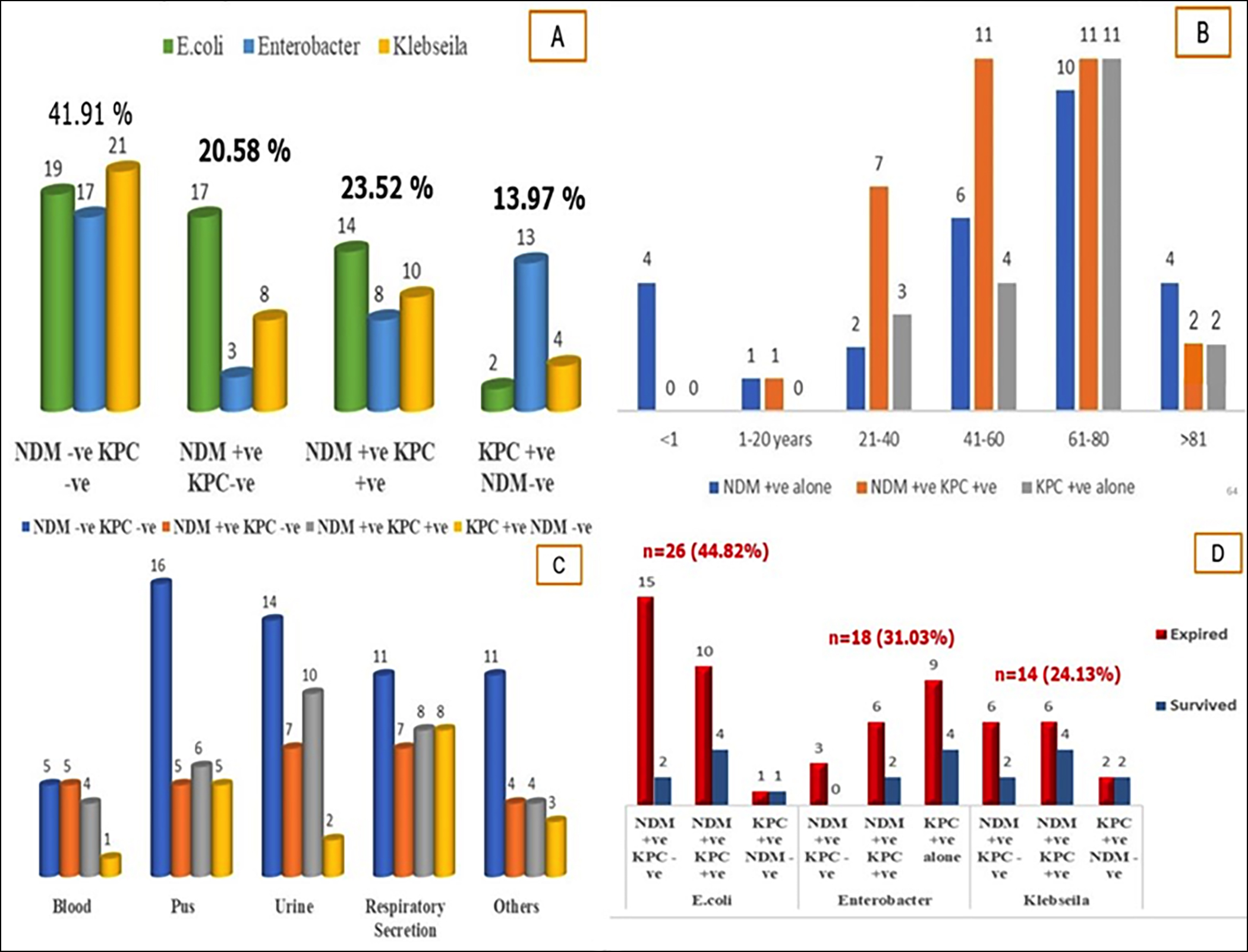 Figure 2: Distribution of New Delhi Metallo-Beta-Lactamases and Klebsiella Pneumoniae Carbapenemases resistant genes in 136 CRE Isolates
Figure 2: Distribution of New Delhi Metallo-Beta-Lactamases and Klebsiella Pneumoniae Carbapenemases resistant genes in 136 CRE Isolates
Frequency of NDM and KPC in different CRE organisms. B) Frequency of NDM and KPC in different age groups. C) Frequency of NDM / KPC genes in different clinical samples. D) Clinical outcome in relation to resistant genes in CRE.
|
Antimicrobials |
Carbapenem Resistant Enterobacterales |
Total (n=136) |
||
|
Escherichia coli (n=52) |
Klebsiella species (n=43) |
Enterobacter species (n=41) |
||
|
Amikacin |
12 (23.07%) |
38 (88.37%) |
36 (87.8%) |
86 (63.23%) |
|
Chloramphenicol |
34 (65.4%) |
24 (58.8%) |
22 (53.65%) |
80 (58.82%) |
|
Ciprofloxacin |
52 (100%) |
42 (97.67%) |
40 (97.56%) |
134 (98.52%) |
|
Co-trimoxazole |
51 (98.07%) |
41 (95.34%) |
39 (95.12%) |
131(96.32%) |
|
Gentamicin |
18 (34.6%) |
41 (95.34%) |
39 (95.12%) |
98 (72.05%) |
Table II: Distribution of resistant genes in relation to gender and organisms.
|
Organism |
Gender |
NDM -ve KPC –ve |
NDM +ve KPC -ve |
NDM +ve KPC +ve |
NDM -ve KPC +ve |
p value |
|
E.coli |
Male |
13 |
10 |
8 |
2 |
0.62 |
|
|
Female |
6 |
7 |
6 |
0 |
|
|
Enterobacter |
Male |
10 |
1 |
4 |
9 |
0.65 |
|
|
Female |
7 |
2 |
4 |
4 |
|
|
Klebsiella |
Male |
10 |
3 |
6 |
3 |
0.58 |
|
|
Female |
11 |
5 |
4 |
1 |
The mean age of adults (>18 years) was 58.65 ± 18.7 years while 4 of the cases were infants less than 1 year of age. The distribution of NDM and KPC is shown in different age groups in Figure 2(B). The majority of the patients (n=32) were between 61-80 years of age. The highest frequency of coexistent KPC and NDM was observed in a group between 41-60 years of age. The frequency of NDM and KPC in relation to gender is depicted in Table II.
According to the clinical samples, the coexistence of KPC and NDM resistance genes was seen mostly in urine samples (n=10/32) followed by respiratory tract samples (n=8/32) and pus (n=6/32). The clinical isolates negative for NDM and KPC were observed mostly in pus followed by urine as given in Figure 2. However, no significant association was observed for presence or absence of resistant genes in CRE positive organisms in different clinical sample sources as depicted in Figure 2.
Out of 136 CRE-positive patients, 79 patients were followed up for the clinical outcomes. Out of 79 patients, n= 58 (73.4%) of the patients expired during hospital stay while n=21 (26.6%) patients were discharged. The majority of mortality was seen in patients with E. coli infection (n=26) followed by Enterobacter (n=18). While mortality was recorded in 14 patients with Klebsiella infection. The associated comorbidities in expired patients were mostly seen in patients with urinary tract infection and chronic kidney disease (CKD) (n=19, 36%) followed by respiratory tract infections and wound infections / abscess (19%, each). Twelve percent of the expired patients had associated comorbidities such as abdominal surgery, road traffic accident, and heart-related problems (congestive heart failure or ischemic heart disease).
The associations of clinical outcomes were then analysed with age and resistant genes. Age showed a significant association with the clinical outcomes [OR= 1.9 (95% CI =1.08-3.32, p<0.024)] while the presence of resistant genes did not depict any association with the clinical outcomes [OR= 1.83 (95% CI =0.93-3.6, p<0.077)].
DISCUSSION
The frequency of coexistent blaNDM -1 and blaKPC resistant genes among clinical isolates of Enterobacterales was found to be high (23.5%) in a set of Karachi population indicating the rapid spread of these resistant genes. NDM and KPC are plasmid-borne and lead to the rapid movement of these resistant genes from cell-to-cell through conjugation with other bacterial cells. As a result, it makes a significant contribution to its broad distribution and associated resistance determinants. Although NDMs and KPCs are not the first or only mechanisms of carbapenem resistance, they are noteworthy because they are frequently undetected by regular susceptibility screening and have a high potential for dispersion. In the USA, the percentage of carbapenem-resistant K pneumoniae increased from 9% in 2002 to 18% in 2004, and subsequently to 38% in 2008.8
The first report of a clinical isolate yielding a KPC outside of the United States came from a patient who had been previously hospitalised in New York City and was made in France.9 Secondly, hospital sinks, OT beds, tables etc. are assumed to be known environmental contaminants.10
It is a major concern that the carbapenem resistance in Enterobacterales is growing worldwide. Other than the Indian subcontinent the NDM carbapenem resistance is reported in other parts of the world as well. A study in India reported 12.3% (57/464) of CRE and with molecular characterisation of NDM by PCR which was positive in all the carbapenem-resistant isolates. The E. coli that was isolated from the urology ward showed more NDM variants, while the mortality rate reported was 23% and 25% in patients infected with isolates positive for blaNDM-1 and blaNDM variants, respectively. Due to their widespread distribution in the Indian subcontinent, NDM has rapidly evolved, as indicated by the variety of genotypic characteristics of blaNDM.11 Another study from New Zealand reported the presence of NDM variants in the CRE organisms who travelled from India. Among the NDM-positive isolates all five isolates carried the plasmid-mediated 16S rRNA methylase RmtC gene, while four of the isolates produced a CTX-M-15 extended-spectrum lactamase and/or plasmid-mediated AmpC-lactamase. The presence of NDM in a country with a meagre antibiotic resistance indicates the global dissemination of diverse phenotypic and genotypic characteristics associated with blaNDM.12
According to a research from Nepal, the most common KPC producers obtained from urine samples were E. coli (57.8 percent), followed by 10.5 percent K. pneumoniae, which is consistent with this study findings.13 The type of patient samples, inpatient wards, and study region can all play a role in the differences between the findings of various studies in terms of the type of common species.
The new resistant infections limit antibiotic alternatives, challenging clinicians for substantial treatment options.11 In this study, all isolates of CRE were sensitive (100%) to Colistin, Tigecycline and Fosfomycin. Sharahi et al. also found Colistin and tigecycline the most efficient antibacterial drugs with 90.3% sensitivity for colistin in clinical isolates.14 Tigecycline has shown promising in vitro activity against CRE and it belongs to the Glycylcycline class of antibiotics. Tigecycline binds to the 30S ribosome of bacteria, restricting t-RNA from entering cells. This subsequently blocks amino acid incorporation into peptide chains, halting bacterial growth and thus being bacteriostatic in nature. The presence of an N,N,-dimethylglycylamido group at position 9 enhances tigecycline's affinity for the ribosomal target by up to five times leading to a greater range of activity and lesser susceptibility to resistance development. Several clinical studies have investigated the efficacy of tigecycline in treating CRE infections, yet these have yielded variable results. However, Colistin and Fosfomycin are bactericidal by action and act in synergy. They impair the Lipopolysaccharide layer by displacing the divalent cations of calcium (Ca2+) and magnesium (Mg2+). This then causes an expansion of the external outer membrane monolayer and inserts its hydrophobic terminal acyl fat chain. As a result, the outer membrane becomes permeable and allows these antibiotics to get inside.15
Overall, in the present study, the most frequent organism observed was E. coli followed by Klebsiella and Enterobacter. Related research performed in Cambodia stated that E. coli (63.9%) was the most prevalent isolate relative to K. pneumoniae (19.8%), with most of the study's samples being urine samples and E. coli was the more frequent source of mild urinary tract infection.16 Urine, in accordance with the above study, was also the most common sample in this study. An overall 6.5% frequency of CRE was found in this study samples. The antibiotic resistance is reported to be increasing every year among clinical isolates worldwide. However, an Indian study reported a gradual increase in CRE from 0-8% from 2006 to 2009, whereas, 5% prevalence was reported in Taiwan.17
In previous investigations, CRE incidences were found to be 2.93 per 100,000 population in the United States and 1.3 per 10,000 hospital admissions in Europe.18 CRE bacteremia has resulted in fatality rates ranging from 20% to 70%, with higher mortality risks attributable to the presence of comorbidities.19 A research in China based on retrospective research from multiple healthcare centres demonstrated an overall CRE infection incidence rate of 4.0 per 10,000 discharges.20 An Egyptian study reported the incidence of CRE in hospital-associated infections (3.7/10,000 patient-days).21 The existence of comorbidities and higher mortality has also been observed in the current study. Earlier in 2011, a study conducted at Pakistani military hospitals laboratories showed CRE carriage rates of 18.5% (n=200) in stool samples, similarly, the carriage rate was found 8.6% and 18.3% in two laboratory-based studies among diarrhoea patients.22,23
A recent study, based on evaluating healthcare facility sink drains reported presence of CRE in 64% of sinks emphasising these sinks can serve as undetected reservoirs for carbapenem-resistant Enterobacterales.10 These figures signify that they are the most prevalent cause of both community and hospital-acquired infections.
The molecular characterisation revealed 23.5% of coexistence of resistant genes i.e., NDM and KPC among the study samples. The co-existence of these two unrelated Carbapenemases renders the isolates highly resistant against many antimicrobials especially carbapenem group of antibiotics. Coexistence of the resistant genes in 2 samples of Klebsiella was reported in Pakistan as well as presence of blaOXA along with these two resistant genes in Pakistan.24
NDM and KPC are Carbapenemases that are present in CRE. Because of the coexistence of various resistance mechanisms, blaNDM positive bacteria as well as blaKPC positive bacteria are frequently resistant to most antimicrobial drugs in addition to β-lactams. Such resistant strains have been identified as the primary source of infections linked with high mortality worldwide, posing substantial clinical care and public health concern. Under these conditions, clinicians rely on a few alternative antibiotics e.g., Colistin, Fosfomycin, and Tigecycline to treat infections caused by CRE.
Further measures need to be taken for detection of isolates producing and coproducing Carbapenemases and can be implemented in regular clinical microbiology laboratories. Although, Colistin, Fosfomycin, and Tigecycline showed very good in vitro antimicrobial sensitivities results in this study but the growing emergence of the powerful resistance mechanisms is a cause for great concern as treatment options are virtually exhausted.
This study will be helpful in increasing awareness among medical professionals, the scientific community, and policymakers about the recent trend in antimicrobial resistance and the need for solutions by restricting the use of antimicrobials and switching to alternative options like screening for resistant genes as routine laboratory investigation. Data from this analysis can be used to guide how and where these strategies can be most efficiently implemented in Pakistan.
This work has certain limitations. No rectal swabs or faecal samples were included in this study which may have resulted in the underestimation of CRE carriage. Whole genome sequencing might have helped in identifying strains. Information on clinical characteristics and outcomes could not be completely acquired because of the limited data and follow-up.
CONCLUSION
An increasing frequency rate of NDM and KPC resistant genes among patients in a private hospital of Karachi was observed. Dissemination of Enterobacterales with co-occurrence of multi-drug resistance genes is alarming. The spread of such strains should be put on foothold by active surveillance, screening of resistant genes and stopping irrational use of antibiotics.
ACKNOWLEDGEMENTS:
No sources of funding were used to assist in the preparation of this manuscript. The authors have no conflicts of interest that are directly relevant to the content of this manuscript.
ETHICAL APPROVAL:
The ethical approval was taken from the Ethical committee of Ziauddin University prior to the sampling.
PATIENTS’ CONSENT:
Informed consent were obtained either from the patients or their close relatives in all the cases.
COMPETING INTEREST:
The authors declared no competing interest.
AUTHORS’ CONTRIBUTION:
FIA: Conceived the project, collected the samples, conducted the lab work and finalised the manuscript.
AS: Performed statistical analysis, interpretation of results, and drafted the manuscript.
RK: Performed lab work and statistical analysis.
SB: Supervised the study along with the finalisation of the manuscript and critically analysed the manuscript.
SAAZ: Collected the samples, lab work and results finalisation.
QJ: Supervised the work and read and finalised the manuscript.
All the authors have approved the final version of the manuscript to be published.
REFERENCES
- Oliveira J, Reygaert WC. Gram negative bacteria PubMed. Treasure Island (FL): Stat pearls publishing; 2020. Available from: Pubmed.ncbi.nlm.nih.gov/30855801.
- Karaiskos I, Giamarellou H. Carbapenem-Sparing Strategies for ESBL producers: When and How. Antibiotics 2020; 9(2):61. doi: 10.3390/antibiotics9020061.
- Queenan AM, Bush K. Carbapenemases: The versatile β-Lactamases. Clinical Microbiology Reviews 2007; 20(3): 440–58. DOI: 10.1128/CMR.00001-07.
- Kumarasamy KK, Toleman MA, Walsh TR, Bagaria J, Butt F, Balakrishnan R, et al. Emergence of a new antibiotic resistance mechanism in India, Pakistan, and the UK: A molecular, biological, and epidemiological study. Lancet Infectious Diseases 2010; 10(9):597–602. doi: 10.1016/S1473- 3099(10)70143-2.
- Paterson D, Doi Y. Carbapenemase-producing enterobacteriaceae. Seminars in Respiratory Critical Care Medicine 2015; 36(01):074–84. doi: 10.1055/s-0035-1544208.
- Procop GW, Koneman EW, Winn WC. Koneman’s color atlas and textbook of diagnostic microbiology. 6th ed. Philadelphia: Lippincott Williams & Wilkins; 2017.
- CLSI (2021). Performance standards for antimicrobial susceptibility testing; Thirty first informational supplement. CLSI document M100-S31. Wayne, PA: Clinical and Laboratory Standards Institute 2021.
- Arnold RS, Thom KA, Sharma S, Phillips M, Kristie Johnson J, Morgan DJ. Emergence of Klebsiella pneumoniae carbapenemase-producing bacteria. Southern Medical J 2011; 104(1):40–5. doi: 10.1097/SMJ.0b013e3181fd7d5a.
- Naas T, Nordmann P, Vedel G, Poyart C. Plasmid-mediated carbapenem-hydrolyzing-lactamase KPC in a Klebsiella pneumoniae Isolate from France. Antimicrobial Agents Chemotherapy 2005; 49(10):4423–4. doi: 10.1128/AAC. 49.10.4423-4424.2005.
- Apanga PA, Ahmed J, Tanner W, Starcevich K, Van Derslice JA, Rehman U, et al. Carbapenem-resistant Enterobacteriaceae in sink drains of 40 healthcare facilities in Sindh, Pakistan: A cross-sectional study. PLOS ONE 2022; 17(2):e0263297. doi: 10.1371/journal.pone.0263297.
- Rahman M, Shukla SK, Prasad KN, Ovejero CM, Pati BK, Tripathi A, et al. Prevalence and molecular characterisation of New Delhi metallo-β-lactamases NDM -1, NDM -5, NDM -6 and NDM -7 in multidrug-resistant Enterobacteriaceae from India. International J Antimicrobial Agents 2014; 44(1):30-7. doi: 10.1016/j.ijantimicag.2014.03.003.
- Williamson DA, Sidjabat HE, Freeman JT, Roberts SA, Silvey A, Woodhouse R, et al. Identification and molecular characterisation of New Delhi metallo-β-lactamase-1 (NDM -1)- and NDM -6-producing Enterobacteriaceae from New Zealand Hospitals. International J Antimicrobial Agents 2012; 39(6):529-33. doi: 10.1016/j.ijantimicag.2012.02.017.
- Dhungana K, Krishna Awal B, Dhungel B, Sharma S, Raj Banjara M, Raj Rijal K. Detection of Klebsiella pneumoniae Carbapenemase (KPC) and metallo-beta lactamase (MBL) producing gram negative bacteria isolated from different clinical samples in a transplant center, Kathmandu, Nepal. Acta Scientific Microbiology 2019; 2(12):60-9. doi: 10. 31080/ASMI.2019.02.0432.
- Sharahi JY, Hashemi A, Ardebili A, Davoudabadi S. Molecular characteristics of antibiotic-resistant Escherichia coli and Klebsiella pneumoniae strains isolated from hospitalized patients in Tehran, Iran. Annals Clinical Microbiology Antimicrobials 2021; 20(1). Doi.10.1186/s12941-021-00437-8.
- Bolla J-M, Alibert-Franco S, Handzlik J, Chevalier J, Mahamoud A, Boyer G, et al. Strategies for bypassing the membrane barrier in multidrug resistant Gram-negative bacteria. FEBS Letters 2011; 585(11):1682-90. doi: 10. 1016/j.febslet.2011.04.054.
- Caron Y, Chheang R, Puthea N, Soda M, Boyer S, Tarantola A, et al. Beta-lactam resistance among Enterobacteriaceae in Cambodia: The four-year itch. International J Infectious Diseases 2018; 66:74–9. DOI:10.1016/j.ijid.2017.10.025.
- Kang H, Zheng W, Kong Z, Jiang F, Gu B, Ma P, et al. Disease burden and molecular epidemiology of carbapenem-resistant Klebsiella pneumoniae infection in a tertiary hospital in China. Annals Translational Medicine 2020; 8(9):605. doi: 10.21037/atm.2020.03.122.
- David S, Reuter S, Harris SR, Glasner C, Feltwell T, Argimon S, et al. Epidemic of carbapenem-resistant Klebsiella pneumoniae in Europe is driven by nosocomial spread. Nature Microbiology 2019; 4(11):1919-29. doi: 10.1038/s41564- 019-0492-8.
- Kang H, Tamma PD, Simner PJ. Phenotypic detection of carbapenemase-producing organisms from clinical isolates. Kraft CS, editor. J Clinical Microbiology 2018; 56(11). doi: 10.1128/JCM.01140-18
- Han R, Shi Q, Wu S, Yin D, Peng M, Dong D, et al. Dissemination of carbapenemases (KPC, NDM, OXA-48, IMP, and VIM) Among carbapenem-resistant Enterobacteriaceae Isolated from adult and children patients in China. Frontiers Cellular Infection Microbiology 2020; 10. doi: 10.3389/fcimb.2020. 00314.
- Kotb S, Lyman M, Ismail G, Abd El Fattah M, Girgis SA, Etman A, et al. Epidemiology of Carbapenem-resistant Enterobacteriaceae in Egyptian intensive care units using national healthcare–associated infections surveillance data, 2011-2017. Antimicrobial Resistance Infection Control 2020; 9(1). doi: 10.1186/s13756-019-0639-7.
- Perry JD, Naqvi SH, Mirza IA, Alizai SA, Hussain A, Ghirardi S, et al. Prevalence of faecal carriage of Enterobacteriaceae with NDM -1 carbapenemase at military hospitals in Pakistan, and evaluation of two chromogenic media. J Antimicrobial Chemotherapy 2011; 66(10):2288-94. doi: 10.1093/jac/dkr299.
- Iqra J, Aizza Z, Muhammad UQ, Hasan E, Junaid A, Abdul W. Multi-drug resistant Klebsiella pneumoniae causing urinary tract infections in children in Pakistan. African J Microbiology Research 2014; 8(4):316–9. Doi: 10.5897/AJMR2013.6409.
- Khaliq A, Rahman H, Khaliq H, Khan Z, Khan TA, Sheer R, et al. blaKPC, blaNDM and blaOXA-48 Producing Klebsiella pneumoniae: First report from Northwest Pakistan. J Pharmacy Pharmacology 2020; 9(1). doi.org/10.17265/2328-2150/ 2021.01.001.