Identification and Association of Polymorphism rs2073618 of the Osteoprotegerin Gene in Type 2 Diabetics with and without Retinopathy
By Erum Tariq, Palvasha Waheed, Asifa Majeed, Amir RashidAffiliations
doi: 10.29271/jcpsp.2023.09.959ABSTRACT
Objective: To identify and determine the association of SNP (rs2073618) of OPG gene in diabetics with and without retinopathy and in healthy controls.
Study Design: Descriptive study.
Place and Duration of the Study: Department of Biochemistry and Molecular Biology, Army Medical College, Rawalpindi in collaboration with Chemical Pathology Laboratory, Pak Emirates Military Hospital, Rawalpindi and Armed Forces Institute of Ophthalmology, Rawalpindi, from June 2021 to May 2022.
Methodology: Participants aged 25-70 years were inducted and divided into three equal groups. Group I consisted of diabetics with retinopathy (n = 50), group II was diabetics without retinopathy (n = 50), and group III was healthy individuals (n = 50). DNA was extracted and allele specific PCR technique was adopted using specifically designed primers. Results were analysed using the software Statistical Package for Social Sciences (SPSS) version 22.0 and online bio-informatics tool SNPstats.
Results: CC, CG, and GG genotypes were found to be present in 94%, 4%, and 2% in diabetics without retinopathy, 92%, 4%, and 4% in diabetics with retinopathy, respectively, and 100% presence of CC genotype only in healthy controls. C and G alleles were present in 96% and 4%, respectively, in diabetics without retinopathy, with 100% presence of only C allele in healthy subjects. The genotypic assessment using the models showed no significant association.
Conclusion: SNP rs2073618 of OPG gene was identified in all study groups without any significant distribution or association with the development of diabetic retinopathy. The major genotype C/C was found in the majority of subjects in all groups.
Key Words: Allele specific PCR, Diabetic retinopathy, Single nucleotide polymorphism, Type 2 Diabetes mellitus.
INTRODUCTION
Diabetes mellitus (DM) is a chronic metabolic disease.1 There are various types of DM on the basis of clinical presentation and aetiology. These include type 1 Diabetes mellitus (T1DM), type 2 Diabetes mellitus (T2DM) and gestational Diabetes mellitus (GDM). Monogenic and secondary diabetes are some of the other less common types of diabetes.1-4 The main cause of T2DM is insulin-resistance. This insulin-resistance occurs due to dysfunction of beta cells.1 Around 90% cases of DM belong to the category of T2DM. In this state, response to insulin is negligible, and thus is called insulin-resistance. T2DM is mostly present in people above the age of 45 years. Due to increase in levels of physical inactivity, energy-dense diet and obesity among the children, younger adults and adolescents, T2DM is seen to be increasing.5,6
One of the major complications of DM is diabetic retinopathy (DR). It is one of the leading causes of blindness in the working population. DR is clinically classified into two stages: Proliferative diabetic retinopathy (PDR) and Non – proliferative diabetic retinopathy (NPDR). NPDR is the early stage of DR. Its main features are the changes in retinal vasculature that may lead to capillary occlusion and increased vascular permeability. The patients of NPDR are mostly asymptomatic but micro-aneurysms, hard exudates and haemorrhages can be detected in these patients via fundus photography. PDR is an advanced stage of DR. Neo-angiogenesis is the main feature of PDR. Vitreous haemorrhage due to bleeding of new abnormal vessels into the vitreous chamber or retinal detachment due to traction may cause severe vision loss in the patients of PDR. The clinical features of the vascular abnormalities of retina lead to the diagnosis of DR.7 The pathogenesis of DR involves angiogenesis.
Osteoprotegerin (OPG) is a newly identified glycoprotein which shows a correlation with the development of angiogenesis.8 OPG protein is secreted under the control of OPG gene. This gene belongs to a member of TNF receptor super-family (TNFRSF11B). It has 5 exons. OPG gene is located at 8q24.12 position on the long arm of chromosome 8. Single nucleotide polymorphism rs20173618 is located in the exon 1 of the OPG gene. OPG is known as an important factor for regulating endothelial cell function, angiogenesis and vasculogenesis. OPG is a member of TNF receptor super-family. Trans-membrane domain is not found in OPG and it is mostly secreted into the extracellular compartment. OPG was initially known for its role in the regulation of bone metabolism by causing paracrine signalling between osteoclasts and osteoblasts. But recently, OPG is found to have a role in the maintenance of endothelial cell function in tumour angiogenesis and vascular diseases. So, it can be concluded that OPG may play a role in the pathogenesis of vascular endothelial cell dysfunction and various other micro-vascular and macro-vascular complications of DM.9
The linking pathway between the higher OPG levels and vascular complications is related to vascular damage and endothelial dysfunction. One of the main mechanism that may lead to the production of OPG may include hyperglyacemia by the activation of inflammatory processes.10
This study was conducted to identify the genotypic and allelic frequency of the polymorphism rs2073618 of the OPG gene that belongs to TNF receptor super-family; to assess its role and association in causing T2DM and DR.
METHODOLOGY
This study was conducted at the Multidisciplinary Lab 1 (CREAM), Department of Biochemistry and Molecular Biology, Army Medical College (AMC), in collaboration with the the Armed Forces Institute of Ophthalmology (AFIO) and Chemical Pathology Laboratory (CPL), Military Hospital (MH), Rawalpindi. It was conducted after the approval from the AMC Ethical Review Board (ERC/ID/110 dated 23.06.2021).
It was a case-control association study, and non-probability convenient sampling technique was employed. The study was completed in one year from June 2021 to May 2022. MAF reported in 10000 genome with frequency 0.3 was taken in account. The calculated sample size was 46 with odd ratio of 2.5 and population disease prevalence 28% considering log-additive genetic model on QUANTO program. The total number of subjects enrolled in the study were 150. All the subjects were equally divided into three groups. Group I consisted of diabetic patients with retinopathy (n = 50), group II consisted of diabetics without retinopathy (n = 50) and group III comprised age and gender matched healthy individuals (n = 50). Consent of all the subjects was taken before the conduct of the study.
All the patients of T2DM with retinopathy of both genders aged between 25-70 years were included in group I, all the patients of T2DM without DR, with any duration of diabetes including the newly diagnosed patients from both genders aged between 25-70 years were included in group II, and all the healthy subjects of both genders aged between 25-70 years were included in group III. All T1DM subjects, all the patients with retinopathy due to causes other than diabetes such as atherosclerosis, hypertension, systemic vasculitis, systemic infections, blood dyscrasias, radiations, etc., patients with ocular diseases other than DR like cataract, glaucoma, papillopathy, and ocular surface diseases such as retinal vein occlusion and retinal macro-aneurysms etc., patients with history of retinal laser therapy or ocular surgeries, and patients with gestational diabetes were excluded.
The medical and surgical history of all patients, along with their demographic data were recorded on the pre-designed proforma. DNA extraction of all the blood samples was done by phenol-chloroform method.11 The agarose gel electrophoresis (1% agarose gel) was used to determine the quality and quantity of extracted DNA. Specific primers were designed using Web-based Allele Specific Primer (WASP) online bioinformatics software. The primers sequence of all the designed primers was as follows:
Wild type forward primer 5' –GGGACCACAATGAACAGC-3'
Mutant forward primer 5' –GGGACCACAATGAACAGG-3'
Common reverse primer 5' –CATGGCATAACTTGAAAGC-3'
Allele-specific polymerase chain reaction (AS-PCR) was performed on the extracted DNA using designed primers to check for the presence of polymorphism rs2073618. The size of the PCR product was 204bp. The composition of PCR mixture was Taq buffer 2.5ul, dNTPs mix (Deoxynucleoside triphosphates) 0.5ul, MgCl2 2ul, forward primer (wild or mutant) 1ul, common reverse primer 1ul, Taq polymerase 0.5ul, DNA template 1.5ul, and NF (nuclease free) water 16ul.
The thermal cycling parameters for AS-PCR were set as hot, starting at 95ºC for 8 minutes, denaturation at 94ºC, annealing at 56.9ºC, and extension at 72ºC for 40 seconds each. Final extension was done at 72ºC for 7 minutes. Total 35 cycles were repeated for the process.
Gel electrophoresis (2% agarose gel) was performed again to identify the polymorphism. Results were analysed using the software Statistical Package for Social Sciences (SPSS) version 22.0. to measure the genotypic and allelic frequencies along with percentages by using the descriptive statistics and their p-value by using the post-hoc Tukey test of the software and online bio-informatics tool SNPstat (https://www.snpstats.net/start.htm) to see the association of genotypes with the disease by adopting different models by applying Chi-square test, Fisher's exact test for Hardy Weinberg equilibrium and logistic regression analysis to measure OR and association studies. The p value equal or less than 0.05 was considered significant (i.e. p ≤0.05).
The selection criteria of SNP was that it was a reported SNP, and many researches have been performed on this SNP in different ethnicities to evaluate its role in causing different complications. However, a study on this SNP to see its role in causing DR and T2DM was done by the authors for the first time in Pakistan.
RESULTS
Allele-specific PCR products of the variant rs2073618 are shown in Figure 1.
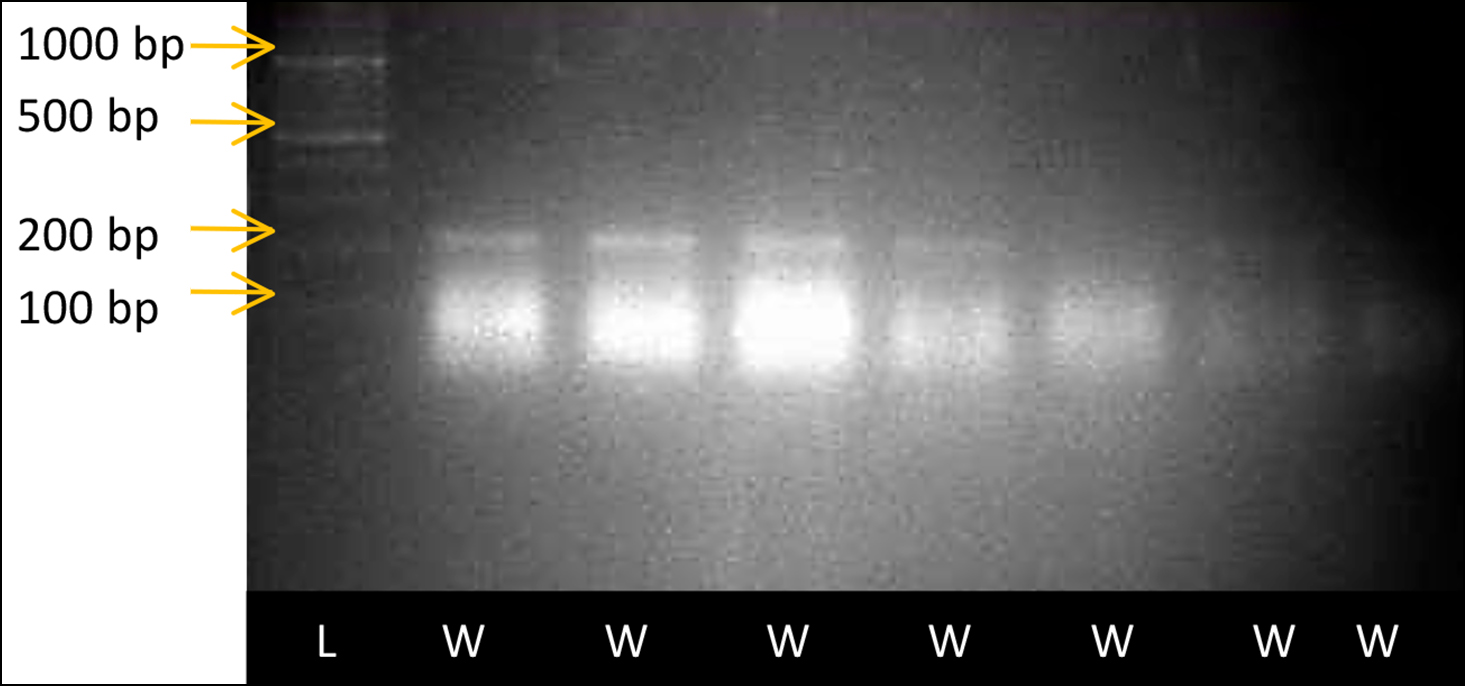 Figure 1: Allele-specific PCR result of single nucleotide variant rs2073618 of OPG gene, as visualised through Gel documentation system. Wells were numbered from 1-8. Well no.1 had DNA ladder of 100bp size. Wild reactions of samples no 105, 106, 107, 108, 109, 110 and 111 were loaded in wells no 2, 3, 4, 5, 6, 7, and 8, respectively. Bands were observed in wells no 2, 3, 4, 5, 6, 7, and 8 with band size 204 bp, indicating wild homozygous genotype C/C of the respective samples.
Figure 1: Allele-specific PCR result of single nucleotide variant rs2073618 of OPG gene, as visualised through Gel documentation system. Wells were numbered from 1-8. Well no.1 had DNA ladder of 100bp size. Wild reactions of samples no 105, 106, 107, 108, 109, 110 and 111 were loaded in wells no 2, 3, 4, 5, 6, 7, and 8, respectively. Bands were observed in wells no 2, 3, 4, 5, 6, 7, and 8 with band size 204 bp, indicating wild homozygous genotype C/C of the respective samples.
When diabetics without retinopathy were compared with healthy controls, the genotypic frequency for CC, CG, and GG genotypes were found to be 94%, 4%, and 2% in cases of diabetics without retinopathy and 100% presence of CC genotype in all healthy subjects. The allelic distribution for C and G allele among the groups were 96% of C allele and 4% G allele present in diabetics without retinopathy and 100% presence of only C allele in all healthy control subjects (Table I). The calculated p-value was 0.424 for all the observed genotypes in these two groups which was not significant.
When SNP rs2073618 association with response status was observed between the diabetics without retinopathy and control group, different models were assessed using the software. The odd ratio (OR) was observed to be 1 or 0 in all the models for all the genotypes. Using the results of AIC values of different models, the model with least AIC value was adopted for the SNP. The least AIC value was observed to be 124.6 in dominant and log-additive model. No association of the SNP rs2073618 in diabetics without retinopathy was found in the selected models as OR was 1.00 in CC genotype, which was not significant and showed no association. In over-dominant gene model, the heterozygote produced by two homozygote parents showed a phenotype that was more pronounced than that of the parents. Over-dominant model showed no association of the SNP as OR was 1.00 in the genotype (Table II).
Genotypic and allelic frequencies along with their percentages were calculated using descriptive statistics of the SPSS software version 22.0 and p-value was calculated using the post hoc Tukey test of the SPSS version 22.0.
Online bio-informatics tool SNPstat (https://www.snpstats.net/ start.htm) was used to see the association of genotypes with the disease by applying Chi-square test, Fisher's exact test for Hardy Weinberg equilibrium and logistic regression analysis.
When diabetics with retinopathy were compared with healthy controls, the genotypic frequencies for CC, CG, and GG genotypes were found to be 92%, 4%, and 4% in cases of diabetics with retinopathy, respectively and 100% presence of CC genotype in all healthy subjects. The allelic distribution for C and G allele among the groups were 94% of C allele and 6% of G allele present in diabetics with retinopathy and 100% presence of only C allele in all healthy control subjects (Table I). The calculated p-value was 0.148 for all the observed genotypes. This is an insignificant value.
When SNP rs2073618 association with response status was observed between the diabetics with retinopathy and control group, different models were assessed using the software. The OR was observed to be 1 or 0 in all the models for all the genotypes. But using the results of AIC values of different models, the model with least AIC value was adopted for our SNP. Least AIC value was observed to be 137.6 in dominant and log-additive model. But no association of the SNP rs2073618 in diabetics with retinopathy was found in the selected models as OR was 1.00 in CC genotype that was not significant and showed no association (Table III).
Online bio-informatics tool SNPstat (https://www.snpstats.net/ start.htm) was used to see the association of genotypes with the disease by applying Chi-square test, Fisher's exact test for Hardy Weinberg equilibrium and logistic regression analysis.
DISCUSSION
The results showed no association among the genotypes and alleles of rs2073618 of the OPG gene in the development of T2DM and DR. The values of genotypic and allelic frequencies and the models adopted using the software showed no significant association of the SNP rs2073618 with the development of disease.
CC genotype was observed as a major genotype in all groups and C allele was observed as a major allele in the groups.
The CC genotype or C allele was found to be present in healthy as well as diseased groups in this study. So no correlation of SNP rs2073618 with the development of DM without retinopathy or DM with retinopathy can be assessed. Many studies have been performed world-wide that either negate or correlate with the findings of this study due to different ethnicities and study populations.
A study was performed on Chinese Han population to investigate the association between the polymorphisms in the OPG gene, and DR. The results showed no significant role of the rs2073618 in the development of disease.12
A meta-analysis was performed in 2021, to check the association of polymorphism of OPG gene and interleukin-6 in the peri-implant disease. The results clearly showed significant relation of the rs2073618 of the OPG gene with the development of peri-implant disease. The results showed that the CC genotype of the rs2073618 increases the risk of peri-implant disease. But there was no significant relationship found between the polymorphism of IL-6, G174C and peri-implant disease.13
Table I: Allelic and genotypic frequencies between DR, DM and controls.
|
Genetic variation |
Group I n = 50 (%) |
Group II (Diabetics without retinopathy) n = 50 (%) |
Group III (Healthy controls) n = 50 (%) |
|
|
Genotype |
CC |
46(92%) |
47(94%) |
50(100%) |
|
CG |
2(4%) |
2(4%) |
- |
|
|
GG |
2(4%) |
1(2%) |
- |
|
|
Allele |
C |
94(94%) |
96(96%) |
100(100%) |
|
G |
6(6%) |
4(4%) |
- |
|
|
p-value |
|
0.148 |
0.424 |
|
Table II: Models between DM without retinopathy and control group.
|
SNP rs2073618 association with response status (n=100, adjusted by sex + age) |
|||||||
|
Model |
Genotype |
Status = DM |
Status = *Co |
OR (95% CI) |
p-value |
AIC |
BIC |
|
Co-dominant |
C/C |
47 (94%) |
50 (100%) |
1.00 |
0.22 |
126.6 |
139.6 |
|
C/G |
2 (4%) |
0 (0%) |
0.00 (0.00-NA) |
||||
|
G/G |
1 (2%) |
0 (0%) |
0.00 (0.00-NA) |
||||
|
Dominant |
C/C |
47 (94%) |
50 (100%) |
1.00 |
0.08 |
124.6 |
135 |
|
C/G-G/G |
3 (6%) |
0 (0%) |
0.00 (0.00-NA) |
||||
|
Recessive |
C/C-C/G |
49 (98%) |
50 (100%) |
1.00 |
0.28 |
126.5 |
136.9 |
|
G/G |
1 (2%) |
0 (0%) |
0.00 (0.00-NA) |
||||
|
Over-dominant |
C/C-G/G |
48 (96%) |
50 (100%) |
1.00 |
0.18 |
125.8 |
136.3 |
|
C/G |
2 (4%) |
0 (0%) |
0.00 (0.00-NA) |
||||
|
Log-additive |
--- |
--- |
--- |
0.00 (0.00-NA) |
0.08 |
124.6 |
135 |
|
*Co = Control group. |
|||||||
Table III: Models between DM with retinopathy and control group.
|
SNP rs2073618 association with response status (n=100, adjusted by sex) |
|||||||
|
Model |
Genotype |
Status=DR |
Status = *Co |
OR (95% CI) |
p-value |
AIC |
BIC |
|
Co-dominant |
C/C |
46 (92%) |
50 (100%) |
1.00 |
0.049 |
139.6 |
150 |
|
C/G |
2 (4%) |
0 (0%) |
0.00 (0.00-NA) |
||||
|
G/G |
2 (4%) |
0 (0%) |
0.00 (0.00-NA) |
||||
|
Dominant |
C/C |
46 (92%) |
50 (100%) |
1.00 |
0.014 |
137.6 |
145.4 |
|
C/G-G/G |
4 (8%) |
0 (0%) |
0.00 (0.00-NA) |
||||
|
Recessive |
C/C-C/G |
48 (96%) |
50 (100%) |
1.00 |
0.074 |
140.4 |
148.2 |
|
G/G |
2 (4%) |
0 (0%) |
0.00 (0.00-NA) |
||||
|
Over-dominant |
C/C-G/G |
48 (96%) |
50 (100%) |
1.00 |
0.098 |
140.8 |
148.6 |
|
C/G |
2 (4%) |
0 (0%) |
0.00 (0.00-NA) |
||||
|
Log-additive |
--- |
--- |
--- |
0.00 (0.00-NA) |
0.014 |
137.6 |
145.4 |
|
*Co = Control group. |
|||||||
In 2020, a study was done to determine the role of OPG gene polymorphism and serum osteoprotegerin in the cardio-vascular disease activity and development of subclinical carotid artery atherosclerosis in the patients of rheumatoid arthritis (RA). Polymerised chain reaction (PCR) was used to identify rs2073618 polymorphism and serum OPG concentrations were measured using other specific techniques. The results clearly showed increased levels of serum OPG in atherosclerotic patients. This increased serum OPG level was highly linked with the increased duration of the disease in such patients. The frequencies of alleles CG, GG and CC among both groups were highly comparable.14 However, in this study, no association between the genotypes of rs2073618 was found.
The levels of OPG were also observed to be raised in some micro-vascular diseases. A study was performed in 2017, to investigate the expression of pro-angiogenic and pro-inflammatory factor, OPG, tumour necrosis factor-related apoptosis-induced ligand (TRAIL), receptor activator of nuclear factor – κB ligand (RANKL) and the receptor RAND in PDR patients. The result showed that the levels of vascular endothelial growth factor (VEGF), OPG and soluble RANK were significantly high in the PDR patients as compared to the healthy subjects.15
This study found no association of rs2073618 with the development of DM and DR but few studies have showed the link of this SNP with the development of micro-vascular complications. A pilot study was performed on 114 subjects with T2DM and diabetic foot (DF) (64 with DR and 50 without DR), to determine the genetic predictors of DR among the patients of T2DM and DF. The results clearly showed the correlation of rs759853, rs3134069, and rs2073618 with the development of disease. A few carriers of the C allele of rs2073618 variant were found in the DR patients in the dominant model.16
The limitations of this study were small sample size and restricted funding. Since this study was based on a single institute, there is a need for multi-centre trials along with provision of proper funding and time to validate the findings of this study.
CONCLUSION
The SNP rs2073618 of the OPG gene was identified in all study groups without any significant distribution in diabetics with or without retinopathy. The major genotype C/C was found in the majority of subjects in all the groups.
ACKNOWLEDGEMENT:
The author is thankful to the Department of Biochemistry and Molecular Biology, Army Medical College, Rawalpindi, and the National University of Medical Sciences (NUMS), Rawalpindi.
FUNDING:
A research grant was provided by the National University of Medical Sciences (NUMS), Rawalpindi.
ETHICAL APPROVAL:
Ethical approval was taken from the Ethical Review Committee of Army Medical College / NUMS, Rawalpindi (ERC/ID/110) dated 23.06.2021, prior to the initiation of research work.
PATIENTS’ CONSENT:
Informed and written consent were taken from all the subjects prior to the initiation of research work.
COMPETING INTEREST:
The authors declared no competing interest.
AUTHORS’ CONTRIBUTION:
ET: Data collection, data analysis, and manuscript drafting.
PW, AR: Data interpretation and manuscript revision.
AM: Data analysis and manuscript drafting.
All authors have approved the final version of the manuscript to be published.
REFERENCES
- Goyal R, Jialal I. Diabetes Mellitus type 2. Treasure Island (FL): StatPearls Publishing; 2021.
- Malek R, Hannat S, Nechadi A, Mekideche FZ, Kaabeche M. Diabetes and Ramadan: A multicenter study in Algerian population. Diabetes Res ClinPract 2019; 150:322-30. doi: 10.1016/j.diabres.2019.02.008.
- Picke A-K, Campbell G, Napoli N, Hofbauer LC, Rauner M. Update on the impact of type 2 Diabetes mellitus on bone metabolism and material properties. Endocr Connect 2019; 8(3):R55-R70. doi:10.1530/EC-18-0456.
- Carrillo‐Larco RM, Barengo NC, Albitres‐Flores L, Bernabe‐Ortiz A. The risk of mortality among people with type 2 Diabetes in Latin America: A systematic review and meta‐analysis of population‐based cohort studies. Diabetes Metab Res Rev 2019; 35(4):e3139. doi:10.1002/dmrr. 3139.
- Su YJ, Chen TH, Hsu CY, Chiu WT, Lin YS, Chi CC. Safety of metformin in psoriasis patients with diabetes mellitus: A 17-year population-based real-world cohort study. J Clin Endocrinol Metab 2019; 104(8):3279-86. doi:10.1210/ jc.2018-02526.
- Choi SE, Berkowitz SA, Yudkin JS, Naci H, Basu S. Personalizing second-line type 2 diabetes treatment selection: Combining network meta-analysis, individualized risk, and patient preferences for unified decision support. Med Decis Making 2019; 39(3):239-52. doi: 10.1177/0272989X1982 9735.
- Wang W, Lo AC. Diabetic retinopathy: pathophysiology and treatments. Int J Mol Sci 2018; 19(6):1816. doi:10.3390/ijms19061816.
- Yu G, Ji X, Jin J, Bu S. Association of serum and vitreous concentrations of osteoprotegerin with diabetic retinopathy. Ann Clin Biochemistry 2015; 52(2):232-6. doi: 10.1177/0004563214533669.
- El-Asrar AMA, Struyf S, Mohammad G, Gouwy M, Rytinx P, Siddiquei MM, et al. Osteoprotegerin is a new regulator of inflammation and angiogenesis in proliferative diabetic retinopathy. Investigative Ophthalmol Visual Sci 2017; 58(7):3189-201. doi: 10.1167/iovs.16-20993.
- Grzelka A, Naskręt D, Araszkiewicz A, Uruska A, Wegner M, Zozulińska-Ziółkiewicz D. Higher concentrations of osteoprotegerin in type 1 diabetic patients are related to retinopathy: Results from the poznań prospective study. Adv Clin Exp Med 2017; 26(9):1343-9. doi: 10.17219/acem/65072.
- Sambrook J, Russell DW. Purification of nucleic acids by extraction with phenol: chloroform. Cold Spring Harb Protoc 2006; 2006(1):pdb.prot4455. doi:10.1101/pdb.prot4455.
- Xu H, Li H, Luo Q, Li Y, Huang G, Lei C, et al. The association of OPG polymorphisms with diabetic retinopathy in Chinese population. Ophthalmic Genet 2021; 42(6): 659-63. doi:10.1080/13816810.2021.1946702.
- Motahari P, Rasi A. The relationship between polymorphism of interleukin-6 and osteoprotgerin genes with dental peri-implant disease: A meta-analysis. Gene Rep 2021; 23: 101133. doi:10.1016/j.genrep.2021.101133.
- Alkady EA, Selim ZI, Sayed SK, Yousef HA, Farrag S, El-Hakeim EH. Association of serum osteoprotegerin and osteoprotegerin gene polymorphism with subclinical carotid artery atherosclerosis and disease activity in rheumatoid arthritis patients. Egypt Rheumatol 2020; 42(3):183-8. doi:10.1016/j.ejr.2020.05.003.
- Abu El-Asrar AM, Ahmad A, Alam K, Bittoun E, Siddiquei MM, Mohammad G, et al. Unbalanced vitreous levels of osteoprotegerin, RANKL, RANK, and TRAIL in proliferative diabetic retinopathy. Ocul Immunol Inflamm 2018; 26(8): 1248-60. doi:10.1080/09273948.2017.1343855.
- Mrozikiewicz-Rakowska B, Łukawska M, Nehring P, Szymański K, Sobczyk-Kopcioł A, Krzyżewska M, et al. Genetic predictors associated with diabetic retinopathy in patients with diabetic foot. Pol Arch Intern Med 2018; 128(1):35-42. doi: 10.20452/pamw.4144.