Concordance of Chromosomal Microarray Analysis in Prenatal Diagnosis of Fetuses with Abnormal Ultrasonographic Soft Markers
By Yan Lu, Caihong Liu, Yan JiAffiliations
doi: 10.29271/jcpsp.2023.03.270ABSTRACT
Objective: To analyse the effect of chromosomal microarray analysis (CMA) in the prenatal diagnosis of fetuses with abnormal ultrasonographic soft markers.
Study Design: Descriptive study.
Place and Duration of the Study: Prenatal Diagnostic Centre, Huizhou Central People’s Hospital, from January 2020 to January 2022.
Methodology: A total of 160 pregnant women with abnormal soft markers in the fetuses on prenatal ultrasonography were selected. Amniotic fluid in the second trimester of pregnancy was extracted for CMA. In addition, karyotype analysis of chromosomal G-banding was carried out to analyse the effect of CMA in prenatal diagnosis.
Results: The detection rate of copy number variants (CNVs) by CMA was higher than that by karyotype analysis, which was not statistically significant (p=0.059). Compared with karyotype detection, CMA detected five additional cases of pathogenic CNVs, all of which were cases of microdeletion and microduplication. VOUS cases detected by CMA were mostly concentrated in NT thickening, among which cases of uncertain significance were the most.
Conclusion: The application of CMA in the prenatal diagnosis of fetuses with abnormal ultrasonographic soft markers can improve the detection rate of pathogenicity. As a prenatal diagnostic method, CMA has high application value and is worthy of clinical promotion.
Key Words: Chromosomal microarray analysis, Abnormal ultrasonographic soft markers, Prenatal diagnosis, Diagnostic efficiency.
INTRODUCTION
Congenital diseases are mainly caused by chromosomal abnormalities, manifesting as various malformations. In severe cases, physical and mental development disorders may occur,1,2 which increase the physical and mental burden of the family. At present, ultrasonography is the main clinical method for diagnosing fetal malformations. Ultrasonography can detect fetal structural abnormalities or soft markers that indicate abnormalities. However, the diagnostic efficiency of ultrasonography is limited.
In recent years, chromosomal karyotype analysis and chromosomal microarray analysis (CMA) are widely applied in prenatal diagnosis,3 and chromosome karyotype analysis, as the gold standard of prenatal diagnosis, is widely used in clinical practice. However, this method has a long detection cycle, low resolution, and cannot detect the variation of short chromosome fragments.
As a molecular diagnostic technique, CMA is relatively novel, and can detect chromosomal microdeletions and microduplications compared with karyotype analysis.4 Currently, CMA is a new genetic detection technology that has emerged in recent years. It scan the entire genome, and detect copy variations in chromosome regions, especially minor deletions and duplications, in prenatal detection.
At present, there are few clinical studies on the combination of the two methods for prenatal testing, which is difficult to form effective evidence-based medical evidence. This study aimed to analyse the diagnostic efficiency and clinical significance of CMA in the prenatal diagnosis of fetuses with abnormal ultrasonographic soft markers, and to provide basis for better prenatal genetic counselling and prognosis evaluation of fetus.
METHODOLOGY
The study was approved by the Institutional Ethics Committee of Huizhou Central People’s Hospital. It was conducted after written informed consent was obtained from all the participants. Those with abnormal ultrasound soft markers, single pregnancy, signed informed consent, and complete clinical data were included in this study, and those who had ultrasonically diagnosed fetal structural abnormalities, a history of noninvasive DNA screening, confirmed contact to obvious teratogenic substances, complicated malignant tumours and abnormal coagulation function, and lost in follow-up were excluded. All included pregnant women underwent ultrasonographic soft marker detection, including choroid plexus cyst, nuchal translucency (NT) thickness, short femur, nasal bone hypoplasia or absence, single umbilical artery, and abnormal lateral ventricle width. At 18-24 weeks of pregnancy, 30 mL amniotic fluid was extracted by transabdominal amniocentesis, 10 mL of which was used for CMA. Amniotic fluid (20 mL) was collected for centrifugation at 2,000 rpm for 5 mins. After removing the supernatant, 0.5 mL of amniotic fluid was taken and added into 3 mL culture medium to prepare cell suspension, one week after culture, the bottle was added with 3 mL fresh medium for continuous culture for 1 day, and added into 45 μL 250 ng/mL colchicine to terminate mitosis. Three mL 0.25% digestive trypsin was added, and cell suspension was collected for fixation overnight. After dropping the suspended cells onto glass slides, baking and cooled naturally, and then added into Giemsa solution for staining. After rinsing with running water and drying, the slides were read with fully automatic scanning. Finally, 20 karyotypes with clear bands were selected for karyotype analysis. The amniotic fluid was drawn by abdominal puncture or umbilical vein puncture under the guidance of B-ultrasound. Ten mL amniotic fluid was drawn for the first time from pregnant women less than 24 weeks old, and 10 mL of amniotic fluid and 1 ml umbilical blood were drawn from pregnant women more than 24 weeks old at the first extraction, and the genomic DNA of amniotic fluid was extracted with DNA extraction agent. With reference to the standard process operation of Infinium gene chip detection of Infinium Company, amplify the DNA samples that have been processed into short fragments, obtain the amplified fragments, purify them, then use the fragment enzyme to short fragment them, and conduct biotin labelling; the labelled substance was mixed with the hybridisation solution and denatured, loaded onto a 750k chip for hybridization, scanning and analysis. It was referred to the existing database for interpretation of copy number variation results. According to the relevant criteria of the expert consensus on technical requirements for chromosomal microarray analysis in laboratories.5 The results were included in the OMIM, ISCA, ECAR UCA, NHS UK, GARD, NLM NIH, and PUBMED databases for analysis. Based on the definition of clinical significance, chromosomal copy number variations (CNVs) were classified into benign, pathogenic, and variant of uncertain significance (VOUS). The CNVs with >200-kb deletion and >500-kb duplicated fragment were analysed, while those with <100-kb deletion and <200-kb duplicated fragment rereading were not reported. Karyotype analysis was conducted according to the relevant criteria of An International System for Human Cytogenomic Nomenclature (2016).6 Statistical analysis was carried out using SPSS 19.0. The enumeration data were represented by (n, %) and compared with the chi-square test. The value of p <0.05 was considered statistically significant.
RESULTS
A total of 160 pregnant women treated, in Huizhou Central People’s Hospital from January 2020 to January 2022 were selected, aged between 22-33 years (average, 27.50 ± 5.49 years) and gestational 18-23 weeks (average, 20.92 ± 2.44 weeks).
The detection rate of CNVs by CMA was higher (31.88%) than that by karyotype (22.50%) analysis, without a statistically significant difference (p=0.059). Compared with karyotype analysis, CMA detected 5 additional cases of pathogenic CNVs, all of which were cases of microdeletion and microduplication, as shown in Table I.
CMA detected 33 variants of uncertain significance (VOUS) cases, accounting for 20.63% of 160 fetuses with abnormal ultrasonographic soft markers, and mostly concentrated in NT thickening, as displayed in Table II.
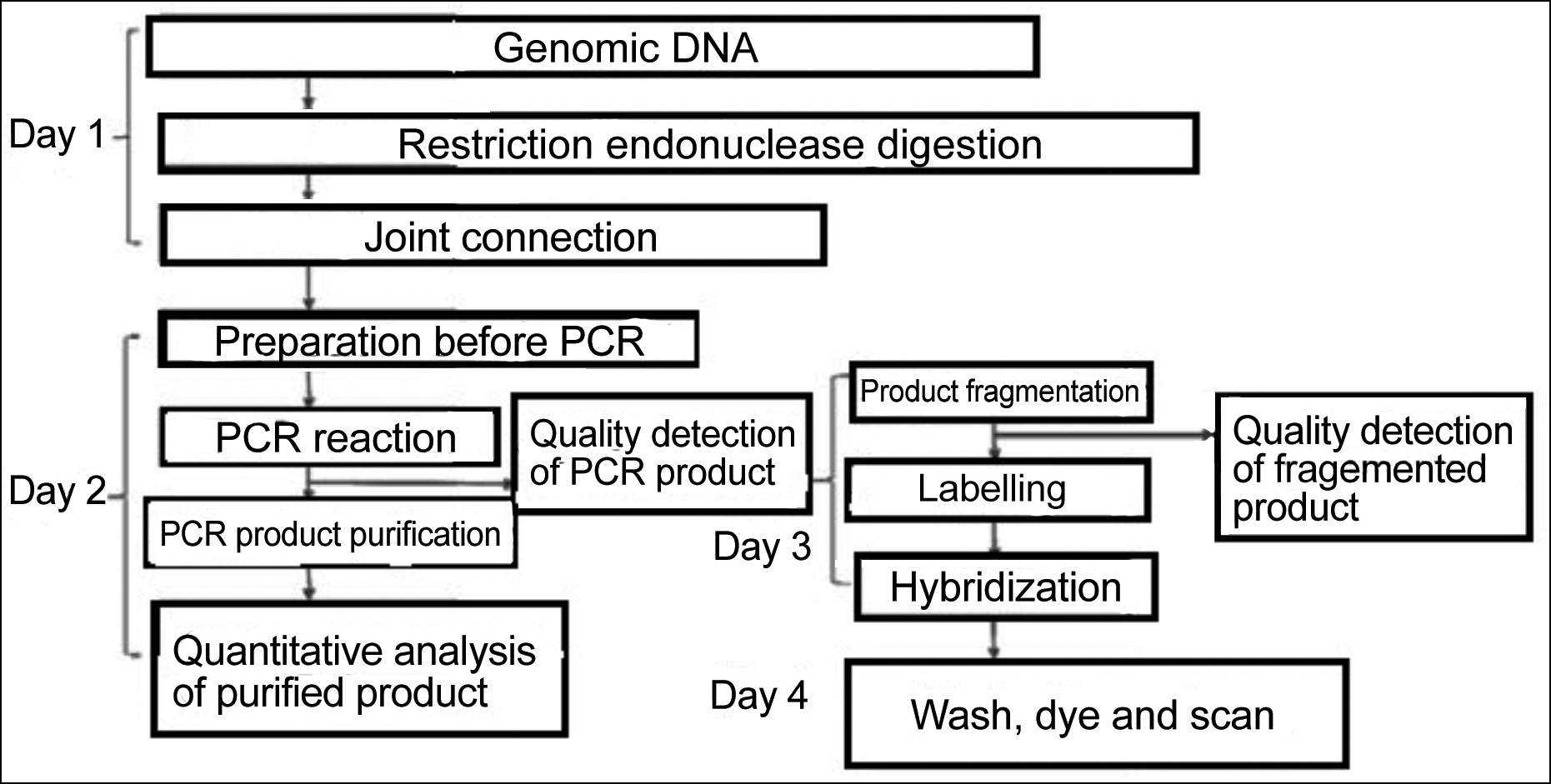 Figure 1: Flowchart of CMA.
Figure 1: Flowchart of CMA.
DISCUSSION
With the continuous progress of ultrasonic technology, fetal structural malformations can be detected early by ultrasonography during pregnancy, which greatly reduces the problems caused by malformed infants to the family and society. Ultrasonographic soft markers are hints of small abnormalities in fetuses, but they do not necessarily mean fetal malformations, and may suggest a normal variation during development, so other means are needed for the confirmation. Chromosomal karyotype analysis and CMA are commonly used in clinical chromosomal diagnosis.7-10 At present, the chromosomal abnormalities are clinically divided into chromosomal copy number abnormalities and chromosomal structure abnormalities, which are caused by abnormal chromosomal position and copy number, thus leading to changes in genetic materials controlled by the abnormal part and manifesting the corresponding chromosomal diseases.11 G-banding karyotype analysis can effectively detect chromosomal polyploid or large fragment changes, but its detection effect of small chromosomal rearrangements is not ideal.12 CMA is usually used for detecting genomic imbalances in fetuses with idiopathic mental retardation, autism, developmental retardation, and a variety of congenital abnormalities.13 CMA for fetuses with abnormal ultrasonographic findings is helpful to exclude congenital diseases caused by chromosomal abnormalities or microdeletion and microduplication.
Table I: Additional detection of pathogenic CMAs.|
Sample No. |
Age |
Gestational weeks |
Ultrasonographic marker |
CMA result |
Reading |
|
1 |
30 |
21 |
NT 4. 2 mm |
22q11.** ×1 (1.39M) |
Microdeletion |
|
2 |
31 |
20 |
NT 3. 8 mm |
16p11.**×1 (0.86M) |
Microdeletion |
|
3 |
34 |
22 |
Ventricular widening |
6q**×1 (2.86M) 17q25.****.*×3 (5.39M) |
Microdeletion Microduplication |
|
4 |
29 |
18 |
Choroid plexus cyst |
2p**.*×3 (3.07M) |
Microduplication |
|
5 |
31 |
19 |
Single umbilical artery |
19p**.*×1 (4.22M) |
Microdeletion |
Table II: Analysis of variants of uncertain significance types detected by CMA (n, %).
|
Soft marker |
NT thickening (n) |
Nasal bone hypoplasia (n) |
Single umbilical artery (n) |
Choroid plexus cyst (n) |
Intraventricular intense spot (n) |
Increased lateral ventricle width (n) |
|
Benign |
1(0.63) |
0(0.00) |
0(0.00) |
0(0.00) |
1(0.63) |
0(0.00) |
|
Pathogenic |
2(1.25) |
0(0.00) |
1(0.63) |
1(0.63) |
0(0.00) |
1(0.63) |
|
Uncertain significance |
12(7.50) |
1(0.63) |
4(2.50) |
8(5.00) |
0(0.00) |
1(0.63) |
In the present study, the detection rate of CMA was higher than the control group, but there was statistically significant difference. Among the additional cases detected by CMA, there were 5 cases of pathogenic CNVs, all of which were caused by microduplication or microdeletion of chromosome segments, and the rest were VOUS cases. The results of 36 cases co-detected by CMA and karyotype analysis were consistent. This may be caused by the difference in the acquisition method for chromosomes. G-banding karyotype analysis is carried out by detecting the karyotypes of the whole chromosome after artificially selecting and culturing cells, while CMA can be performed directly using uncultured amniotic cells, and analyse gene fragments with a large number of known probes to provide genomic information.14 Amniotic cells may cause fluctuations in the number of chromosomal abnormalities detected by karyotype analysis during culture or artificial cell selection. As a microarray analysis, CMA can detect CNVs, the interpretation of which can be divided into benign, uncertain significance, and pathogenic. Pathogenic CNVs are generally believed to occur in functional regions of important genes due to a wide range of chromosomal deletions or rearrangements.15 According to the report by Cheng et al.,16 the diagnostic rate of pathogenic CNVs is 12.2%, and CMA can detect submicroscopic aberrations that cannot be detected by karyotype analysis, with high diagnostic efficiency in prenatal diagnosis of genomic imbalances. CMA can detect chromosomal duplication and deletion with the size of 0.05-0.1 Mb in the genome. However, it still has the limitation that it cannot detect chromosomal structural abnormalities such as balanced translocation and inversion, which still need to be identified by karyotype analysis.17,18 Pylyp et al. has reported that after including the deletion above 0.015 M and duplication below 0.02 M in CMA for fetuses with abnormal ultrasonographic soft markers,19 the additional detection rate is 6.5%, which is similar to the results of this study. According to the statistics published in 2018, it is known that the incidence of microdeletion or microduplication syndromes is 0.04-0.1 %, the independent risk factors do not include maternal age, and the fetuses may suffer from moderate to severe diseases after birth.20 Studies have proved that genome-wide detection will not significantly improve the detection rate of pathogenic CNVs, and the application of CMA in the prenatal evaluation of fetuses with major structural abnormalities has certain advantages.21,22 In this study, VOUS cases detected by CMA were mostly concentrated in NT thickening, accounting for 45.45%. Abnormal NT thickness is an important ultrasonic indication for monitoring fetal chromosomal abnormalities and cardiovascular abnormalities.23 It has been pointed out that when NT is thicker than 3 mm in ultrasonography, CMA should be performed.24 In the VOUS cases, there were 26 cases of uncertain significance, among which 6 were detected after parent CMA, 4 were inherited from one of the parents, 2 were caused by mutations during development, and 5 chose to continue pregnancy. Follow-up hitherto, 2 cases were delivered, 3 cases were still in pregnancy, and the others had no obvious abnormalities. Therefore, the notification of detection results should be more professional to avoid increasing parents’ anxiety, so as to improve unnecessary termination of pregnancy. At present, non-invasive prenatal screening is increasingly widely used, and its operation causes less harm and pain to pregnant women. However, when the fetuses are suspected of having chromosomal abnormalities after screening, puncture is still necessary for karyotype analysis and CMA in time.25 Due to the limitations in the sample size of this study and research time, the application scheme and theory of CMA in prenatal diagnosis of fetuses with abnormal ultrasonographic soft markers need to be further improved.
CONCLUSION
In conclusion, the application of CMA in prenatal diagnosis of fetuses with abnormal ultrasonographic soft markers can improve the detection rate and has a warning effect on cases of microdeletion and microduplication, and it is worthy of clinical promotion.
ETHICAL APPROVAL:
The study was approved by the Institutional Ethics Committee of Huizhou Central People’s Hospital (No. 2021021; date: March 12, 2021).
PATIENTS’ CONSENT:
The author declare that they have obtained from patients informed consent to publish the data concerning this case.
COMPETING INTERESTS:
The authors declare no conflicts of interest.
AUTHORS’ CONTRIBUTION:
YL: Designed the study, and is responsible and accountable for the accuracy or integrity of the work.
CL: Collected and analysed the data.
YJ: Wrote the manuscript.
All the authors have read and agreed to the published version of the manuscript.
REFERENCES
- Hui L, Poulton A, Kluckow E, Lindquist A, Hutchinson B, Pertile MD, et al. A minimum estimate of the prevalence of 22q11 deletion syndrome and other chromosome abnormalities in a combined prenatal and postnatal cohort. Hum Reprod 2020; 35(3):694-704. doi:10.1093/humrep/dez286.
- Iwarsson E, Conner P. Detection rates and residual risk for a postnatal diagnosis of an atypical chromosome aberration following combined first-trimester screening. Prenat Diagn 2020; 40(7):852-9. doi:10.1002/pd.5698.
- Sagi-Dain L, Cohen Vig L, Kahana S, Yacobson S, Tenne T, Agmon-Fishman I, et al. Chromosomal microarray vs. NIPS: analysis of 5541 low-risk pregnancies. Genet Med 2019; 21(11):2462-7. doi:10.1038/s41436-019-0550-x.
- Chung CCY, Chan KYK, Hui PW, Au PKC, Tam WK, Li SKM, et al. Cost-effectiveness analysis of chromosomal microarray as a primary test for prenatal diagnosis in Hong Kong. BMC Pregnancy Childbirth 2020; 20(1):109. doi:10.1186/s12884- 020-2772-y.
- Expert committee of prenatal screening and diagnosis interlaboratory quality evaluation, clinical laboratory center, national health care commission. Expert consensus on technical requirements for chromosome microarray analysis laboratories. Chin J of Lab Med 2019; 42(9):745-51. doi:10.3760/ cma.j.issn.1009-9158.2019.09.006.
- Jean Mc Gowan-Jordan, Ottawa, Ont, Annet Simons. An International System for Human Cytogenomic Nomenclature. Wurzburg Rep of Cytog and Geno Res 2016; 149:1-2.
- Singer A, Maya I, Sukenik-Halevy R, Tenne T, Lev D, Ben Shachar S, et al. Microarray findings in pregnancies with oligohydramnios - a retrospective cohort study and literature review. J Perinat Med 2019; 48(1):53-8. doi:10.1515/jpm-2019-0228.
- Shi Y, Ma J, Xue Y, Wang J, Yu B, Wang T. The assessment of combined karyotype analysis and chromosomal microarray in pregnant women of advanced maternal age: A multicenter study. Ann Transl Med 2019; 7(14):318. doi:10. 21037/atm.2019.06.63.
- Muys J, Blaumeiser B, Jacquemyn Y, Bandelier C, Brison N, Bulk S, et al. The belgian microarray prenatal (BEMAPRE) database: A systematic nationwide repository of fetal genomic aberrations. Prenat Diagn 2018; 38(13):1120- 1128. doi:10.1002/pd.5373.
- Sansović I, Ivankov AM, Bobinec A, Kero M, Barišić I. Chromosomal microarray in clinical diagnosis: A study of 337 patients with congenital anomalies and developmental delays or intellectual disability. Croat Med J 2017; 58(3): 231-8. doi:10.3325/cmj.2017.58.231.
- Andrade FM, Drumond CL, Affonso MN. EP14.40: Abnormal chromosomes 18 on microarray :SNP ‐array analysis with normal band G karyotype in a severe growth ‐restricted fetus. Ultra in Obste & Gyne 2017; 50(S1):326-6. doi: 10. 1002/uog.18556.
- Herghelegiu D, Ionescu CA, Pacu I, Bohiltea R, Herghelegiu C, Vladareanu S. Antenatal diagnosis and prognostic factors of aneurysmal malformation of the vein of Galen: A case report and literature review. Medicine (Baltimore) 2017; 96(30):e7483. doi:10.1097/MD.0000000000007483.
- Bajaj Lall M, Agarwal S, Paliwal P, Saviour P, Joshi A, Joshi A, et al. Prenatal diagnosis by chromosome microarray analysis, an Indian experience. J Obstet Gynaecol India 2021; 71(2):156-67. doi:10.1007/s13224-020-01413-6.
- Hao M, Li L, Zhang H, Li L, Liu R, Yu Y. The difference between karyotype analysis and chromosome microarray for mosaicism of aneuploid chromosomes in prenatal diagnosis. J Clin Lab Anal 2020; 34(12):e23514. doi:10.1002/jcla.23514.
- Souza LCD, Santos APD, Sgardioli IC. Phenotype comparison among individuals with developmental delay/intellectual disability with or without genomic imbalances. J intellect Disabil Res 2019; 63(11): 1379-89. doi: 10.1111/jir.12615.
- Cheng SSW, Chan KYK, Leung KKP, Au PKC, Tam WK, Li SKM, et al. Experience of chromosomal microarray applied in prenatal and postnatal settings in Hong Kong. Am J Med Genet C Semin Med Genet 2019; 181(2):196-207. doi:10. 1002/ajmg.c.31697.
- Levy B, Wapner R. Prenatal diagnosis by chromosomal microarray analysis. Fertil Steril 2018; 109(2):201-12. doi:10.1016/j.fertnstert.2018.01.005.
- Cicatiello R, Pignataro P, Izzo A, Mollo N, Pezone L, Maruotti GM, et al. Chromosomal microarray analysis versus karyotyping in fetuses with increased nuchal translucency. Med Sci (Basel) 2019; 7(3):40. doi:10.3390/medsci7030040.
- Pylyp LY, Spynenko LO, Verhoglyad NV, Mishenko AO, Mykytenko DO, Zukin VD. Chromosomal abnormalities in products of conception of first-trimester miscarriages detected by conventional cytogenetic analysis: A review of 1000 cases. J Assist Reprod Genet 2018; 35(2):265-71. doi:10.1007/s10815-017-1069-1.
- Goldenberg P. An Update on common chromosome microdeletion and microduplication syndromes. Pediatr Ann 2018; 47(5):e198-e203. doi:10.3928/19382359-20180 419-01.
- Egloff M, Hervé B, Quibel T, Jaillard S, Le Bouar G, Uguen K, et al. Diagnostic yield of chromosomal microarray analysis in fetuses with isolated increased nuchal translucency: A French multicenter study. Ultrasound Obstet Gynecol 2018; 52(6):715-21. doi:10.1002/uog.18928.
- Szewczyk K, Bik-Multanowski M. Molecular karyotyping in early miscarriages: Potential for the routine use of cytogenetic microarrays. J Obstet Gynaecol. 2018; 38(4):585-6. doi:10.1080/01443615.2017.1383976.
- Cicatiello R, Pignataro P, Izzo A, Mollo N, Pezone L, Maruotti GM, et al. Chromosomal microarray analysis versus karyotyping in fetuses with increased nuchal translucency. Med Sci (Basel) 2019; 7(3):40. doi:10.3390/medsci7030040.
- Maya I, Yacobson S, Kahana S, Yeshaya J, Tenne T, Agmon-Fishman I. Cut-off value of nuchal translucency as indication for chromosomal microarray analysis. Ultrasound Obstet Gynecol 2017; 50(3):332-5. doi:10.1002/uog.18814.
- Wang J, Chen L, Zhou C, Wang L, Xie H, Xiao Y, et al. Prospective chromosome analysis of 3429 amniocentesis samples in China using copy number variation sequencing. Am J Obstet Gynecol 2018; 219(3):287.e1- 287.e18. doi:10.1016/j.ajog.2018.05.030.