CBX2 Expression in Colorectal Mucosa-adenoma-adenocarcinoma Sequence
By Bangting Wang1, Meina Huang2, Xin Wang2, Yujen Tseng1, Feifei Luo1, Jie Liu1Affiliations
doi: 10.29271/jcpsp.2021.09.1051ABSTRACT
Objective: To investigate the expression of chromobox 2 (CBX2) in colorectal adenoma (CRA) and colorectal cancer (CRC), and analyse its correlation with various clinicopathological parameters.
Study Design: Observational study.
Place and Duration of Study: Pathology Department, Huashan Hospital, Fudan University, from December 2019 to December 2020.
Methodology: The mRNA level of CBX2 in colorectal mucosa, CRA and CRC was evaluated in gene expression profiling interactive analysis (GEPIA) and gene expression omnibus (GEO) dataset, then verified by quantitative real-time PCR (qRT-PCR). CBX2 expression by immunohistochemistry was determined in 122 samples, then its correlation was analysed with various clinicopathological parameters. Diagnostic value of CBX2 was estimated by the receiver operating characteristic curve (ROC). Prognostic value of CBX2 mRNA expression was evaluated via the Kaplan-Meier method in GEPIA.
Results: CBX2 expression rate in CRC (89.8%) was greater than adenoma (37.74%) and mucosa (20%). CBX2 protein levels were highest in adenocarcinoma and lowest in mucosa with intermediate level in adenoma. The area under curve (AUC) of CBX2 was 0.810 and 0.734, respectively, in distinguishing CRA from mucosa and CRC from CRA. CBX2 hyperexpression was not significantly correlated with clinicopathological variables, either in CRA or CRC. Kaplan-Meier survival analysis revealed that CBX2 mRNA hyperexpression was not associated with overall survival (OS) of CRC. Although the disease-free survival (DFS) of high CBX2 patients was shorter than those with low expression, but no obvious significance was found in colon adenocarcinoma (COAD, p=0.052) and rectal adenocarcinoma (READ, p=0.097).
Conclusion: CBX2 expression progressively increased in the sequence of mucosa-adenoma-carcinoma, which may be used as a diagnostic biomarker and therapeutic target for CRA and CRC.
Key Words: CBX2, Colorectal adenoma, Colorectal cancer, Biomarker.
INTRODUCTION
Colorectal cancer (CRC) is the third most common malignant tumor worldwide.1 Approximately 85% of colorectal cancers are thought to evolve from conventional adenomas which is referred to as the adenoma-to-carcinoma sequence.2 The development of CRC is a multistep process, initiated by the formation of a benign polyp with the potential to evolve into a carcinoma in situ, aided by accumulation of additional somatic mutations.3 This process is driven by mutations and epigenetic alterations, which usually takes 10–15 years but can occur more rapidly under certain settings (e.g. lynch syndrome, possibly microsatellite unstable tumors).4
A colorectal polyp is defined as a visible protrusion above the surface of the normal intestinal mucosa. Adenomatous polyps may be flat, sessile, sub-pedunculated (having a very short stalk, or peduncle), or pedunculated.5 Although the histology of conventional tubular adenomas is fairly homogeneous, there is an obvious molecular heterogeneity among them that may determine which adenomas have the highest potential to progress into CRC.6 Some studies have estimated that only 10% of all polyps will progress to CRC and approximately 25% of advanced adenomas will progress to CRC.7 Given the high incidence of CRA and its potential of transforming to adenocarcinoma, identifying new biomarkers to confirm its pathology, predicting the possibility of adenoma to adenocarcinoma transition and providing therapeutic targets for patients are of great importance.
Polycomb group (PcG) proteins, major epigenetic regulators, play important roles in establishing genetic expression profile and lineage commitment.8 PcG proteins are often assembled into two major chromatin-associated complexes, known as polycomb repressive complex (PRC) 1 and 2, which play a vital role in the organisation of chromatin structure.9 Additionally, PcG proteins contribute to cell proliferation, stem cell maintenance and self-renewal.10,11 Chromobox (CBX) member proteins, including CBX1-8, are essential regulators of gene expression and developmental programmes.12 Among them, CBX2 is an important component of the PRC1 complex involved in antiviral innate immunity, differentiation of neuron and gonad, and cell proliferation.13,14 Growing evidence suggests that aberrant expression of CBX family members plays a key role in different cancers.15 As an oncogene, CBX2 is an essential regulator of cancer cell cycle and progression. Some studies have stated that CBX2 is a novel biomarker of cancer because CBX2 expression is higher in breast cancer and hepatocellular carcinoma than in adjacent normal tissues.16,17 High CBX2 expression is associated with poor clinical outcomes in hepatocellular carcinoma. Furthermore, CBX2 knockdown significantly inhibits the proliferation of hepatocellular carcinoma cells and induces cell apoptosis.16
Recently, many studies have dedicated to finding meaningful biomarkers for adenoma and adenocarcinoma over the past decade.18 Although CBX2 is a good therapeutic candidate for many cancers, its expression in colorectal adenoma or adenocarcinoma is still unclear.
The aim of the present study was to investigate the expression of CBX2 in colorectal mucosa, adenoma and adenocarcinoma, and to reveal its associations with various clinicopathological parameters and prognosis.
METHODOLOGY
This observational study was conducted at Pathology Department, Huashan Hospital, Fudan University, from December 2019 to December 2020. Twelve pairs of adenoma and adjacent mucosa were collected from the Endoscopy Centre. Pathological sections of 122 cases of mucosa (n=20), adenoma (n=53) and colorectal cancer (n=49) (2019-2020) were obtained from the Department of Pathology and Endoscopy. Information on age, gender, size and location of the lesions, metastasis and stage were collected from the medical records of the patients.
Total RNA was extracted from adenomas and mucosa using the RNeasy protect mini kit (Qiagen). The GoscriptTM reverse transcription system (promega) was used to reverse transcription of mRNA. Each PCR was carried out in 15 μl volume, using SYBR green master mix (Vazyme) for 15 mins at 95°C for initial denaturing, followed by 40 cycles of 95°C for 30s and 60°C for 1 min in the 7500 fast real-time PCR system (applied biosystems). Primers: CBX2-Forward: GAGTGCATCCTGAGCAAGCG; CBX2-Reverse: CCCAGCT GTTATGTTTGGAGG; GAPDH-Forward: AACGGATTTGGTCGTATTG; GAPDH-Reverse: GGAAGATGGTGATGGGATT.
For morphological analysis, 5 μm adenocarcinoma, adenoma and mucosa sections were prepared for hematoxylin eosin staining. After orderly deparaffinised in a series of xylene baths, and rehydrated using a graded alcohol series, paraffin sections were retrieved in sodium citrate buffer (pH 6.0) via pressure cooker heating for 10 mins.
The CBX2 antibody was (GeneTex, USA) used for IHC at a 1:200 dilution incubated at 4o℃ overnight. Secondary antibody was incubated at room temperature for 60 mins, the horseradish peroxidase solution was incubated for 30 mins and performed with diaminobenzidine, according to manufacturer’s recommendations (GBI, USA) and subsequently counterstained with hematoxylin.
Staining intensity was scored by two independent observers, who were blinded to the patient information. The intensity of staining (IS) was divided into four score ranks: 0 = negative; 1 = weak; 2 = moderate; and 3 = strong, and the percentage of positively stained cells (PS): 0 (<5%), 1 (5-25%), 2 (25-50%), 3 (50-75%) and 4 (75-100%).19 The score of each slide: IS x PS (0 - 12). Staining score was classified into two subgroups (whereby those with scores <6 was classified as low expression, and those with score ≥6 were classified as high expression).
The gene expression profile interactive analysis (GEPIA) (http://gepia.cancer-pku.cn/) tool was used to analyse the CBX2, and to compare the expression level of CBX2 in CRC with adjacent tissues, in order to determine the prognostic value of CBX2 in predicting CRC outcomes. The methylation level of CBX2 was searched at Ualcan (http://ualcan.path.uab.edu/).
SPSS software was used for ROC analysis of CBX2 (version 20.0, SPSS Inc, Chicago, USA). ROC and AUC were utilised to evaluate the predictive value of CBX2 for CRA and CRC.
Chi-square test and Fisher’s Exact test were used to assess correlations between CBX2 protein expression and clinicopathological characteristics. Kaplan-Meier curves and log-rank test were used to present the clinical outcomes, including disease-free survival (DFS) and overall survival (OS). IBM SPSS Statistics version 20.0 Software (SPSS Inc., Chicago, IL, USA) and GraphPad Prism 8.0 software (GraphPad Software Inc, CA, USA) were used for statistical analysis. Data were reported as means ± SD. Student's t test is used to compare the difference between two groups, and one-way ANOVA is used to verify the differences among multiple groups based on single factor. So, the authors used student’s t-test and one-way ANOVA was used to analyse the experimental results. Statistical significance was considered at p <0.05.
RESULTS
First, the expression levels of the CBX2 in colon adenocarcinoma (COAD) and rectum adenocarcinoma (READ) were analysed using GEPIA based on TCGA data, and it was found that CBX2 was highly expressed in COAD and READ (Figures 1a and 1b). Next, hyperexpression of CBX2 regulation by the status of gene promoter methylation was investigated. Ualcan was employed to validate the correlation between DNA methylation level and mRNA expression of CBX2. As expected, the hypomethylation of CBX2 promoter was found in COAD and READ (Figures 1c and 1d).
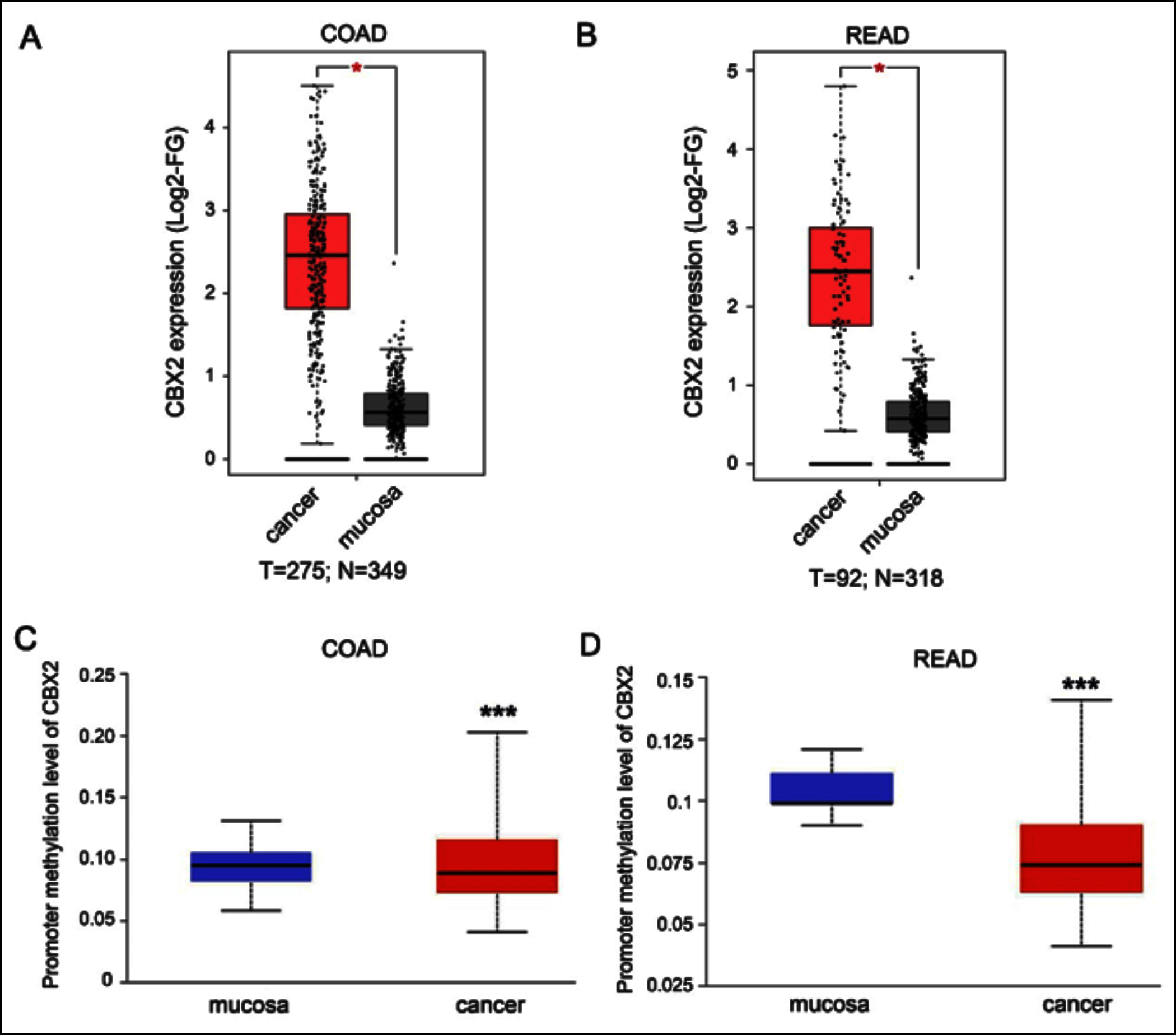 Figure 1: Identifying CBX2 from Ualcan and TCGA database. (A,B) The scatter plot showed that CBX2 was highly expressed in COAD and READ from GEPIA. (C,D). The bar charts showed the methylation level of CBX2 between mucosa and CRC in Ualcan database.
Figure 1: Identifying CBX2 from Ualcan and TCGA database. (A,B) The scatter plot showed that CBX2 was highly expressed in COAD and READ from GEPIA. (C,D). The bar charts showed the methylation level of CBX2 between mucosa and CRC in Ualcan database.
COAD: Colon adenocarcinoma, READ: Rectum adenocarcinoma, Log2-FG: Log2-foldchange. ***Indicates p<0.001, *Indicates p<0.05.
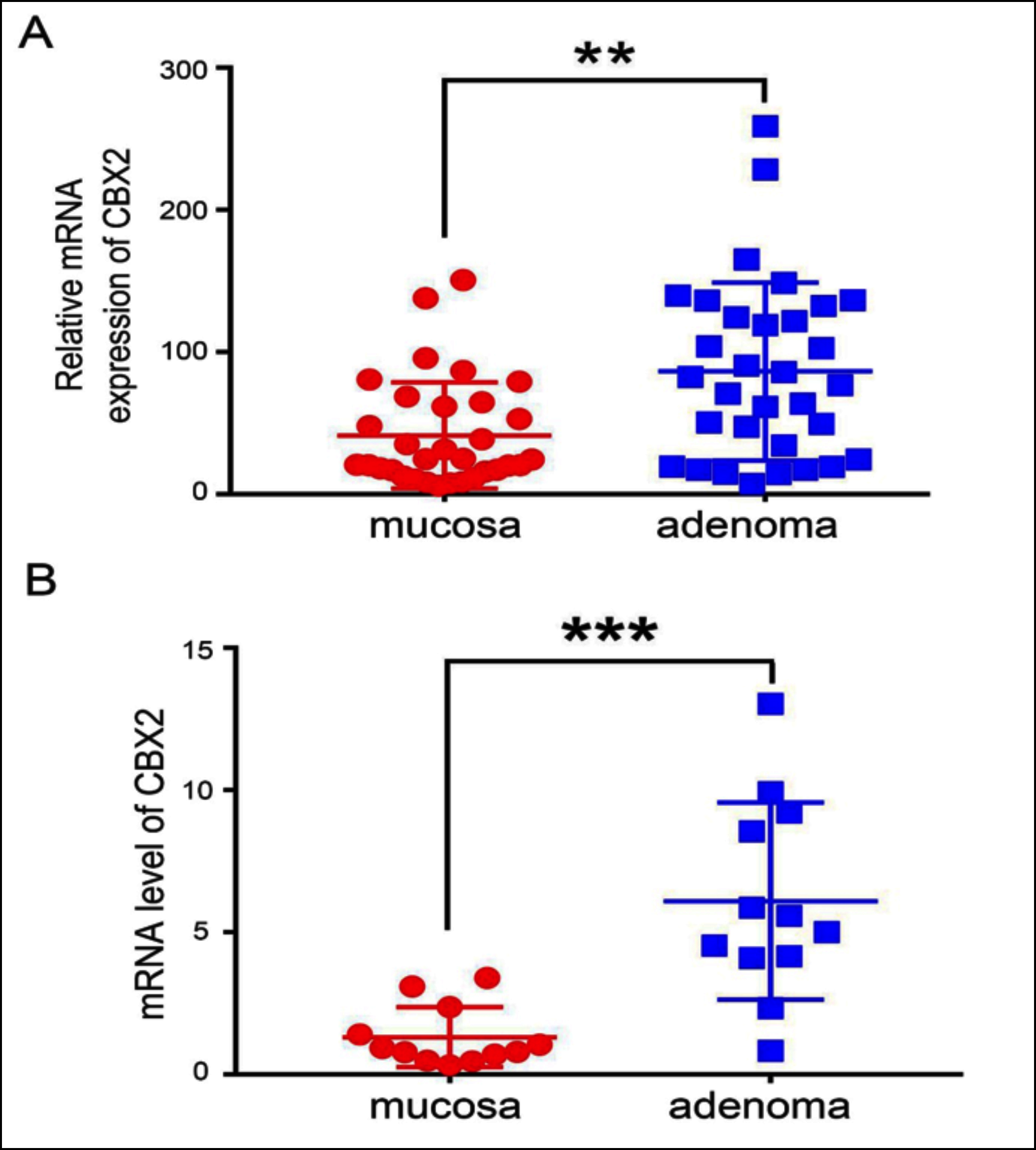 Figure 2: Identifying CBX2 expression in CRA. (A) The relative expression of CBX2 between mucosa and adenoma in GEO database (GEO8671) (p-value <0.01). (B) The mRNA level of CBX2 between mucosa and adenoma by qRT-PCR (p-value <0.001).
Figure 2: Identifying CBX2 expression in CRA. (A) The relative expression of CBX2 between mucosa and adenoma in GEO database (GEO8671) (p-value <0.01). (B) The mRNA level of CBX2 between mucosa and adenoma by qRT-PCR (p-value <0.001).
***Indicates p<0.001, **Indicates p<0.01.
To characterise the expression of CBX2 in adenomas, CRA expression microarray dataset GSE8671 was analysed, including 32 pairs of CRA and adjacent mucosa samples, and compared the relative mRNA expression between mucosa and adenoma. The result showed CBX2 was greatly increased in CRA (Figure 2a). This increased expression in microarray expression profiling was further validated by qRT-PCR, performed on 12 pairs of mucosa and adenoma specimens. The results indicated that the mRNA level of CBX2 was significantly upregulated in CRA (Figure 2b).
The occurrence of CRC followed normal mucosa-adenoma-adenocarcinoma sequence. CBX2 expression was compared in human tissue samples including normal mucosa (n=20), adenoma (n=53) and adenocarcinoma (n=49) by immunohistochemistry. H&E staining exhibited the morphological changes from normal mucosa to adenoma and adenocarcinoma (Figure 3a). Notably, immunohistochemical staining showed an increased trend in protein level of CBX2 in those samples (Figure 3a). Robust CBX2 expression was observed in most of the adenocarcinomas, while adenomas had moderate-signal intensity and normal mucosa showed low or no expression.
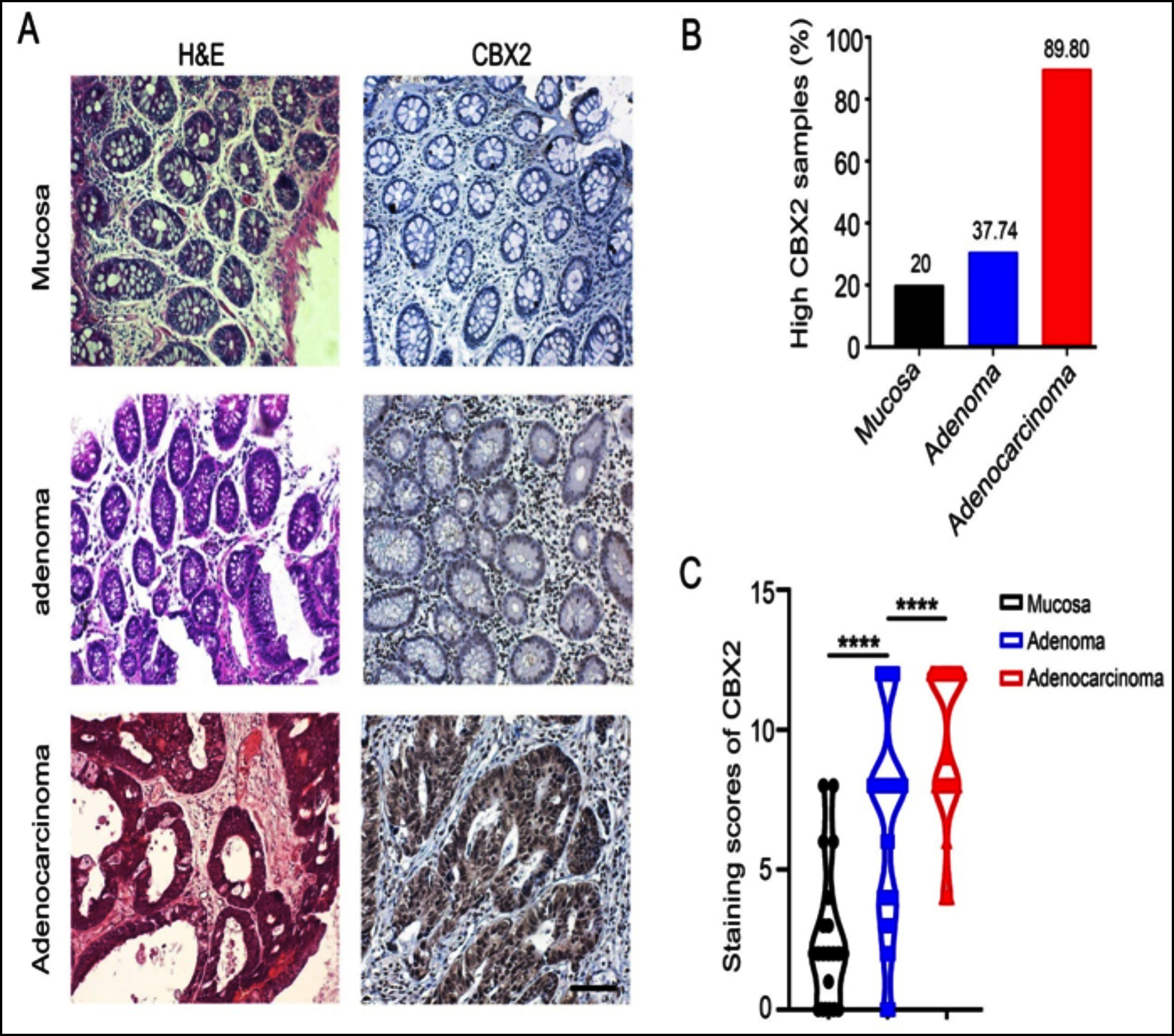 Figure 3: Immunohistochemical staining for the expression of CBX2 in mucosa, adenoma and adenocarcinoma. (A) H&E staining showed the morphologic structures of mucosa, adenoma, and adenocarcinoma; The immunohistochemical staining revealed the expression status of CBX2. (B) Rate of CBX2-hyperexpressionin each histopathological category. (C) The scatter diagram showed immunohistochemical score of CBX2 in mucosa, adenoma and adenocarcinoma (p<0.0001).
Figure 3: Immunohistochemical staining for the expression of CBX2 in mucosa, adenoma and adenocarcinoma. (A) H&E staining showed the morphologic structures of mucosa, adenoma, and adenocarcinoma; The immunohistochemical staining revealed the expression status of CBX2. (B) Rate of CBX2-hyperexpressionin each histopathological category. (C) The scatter diagram showed immunohistochemical score of CBX2 in mucosa, adenoma and adenocarcinoma (p<0.0001).
****Indicates p<0.0001, Black scale bar: 200 μm.
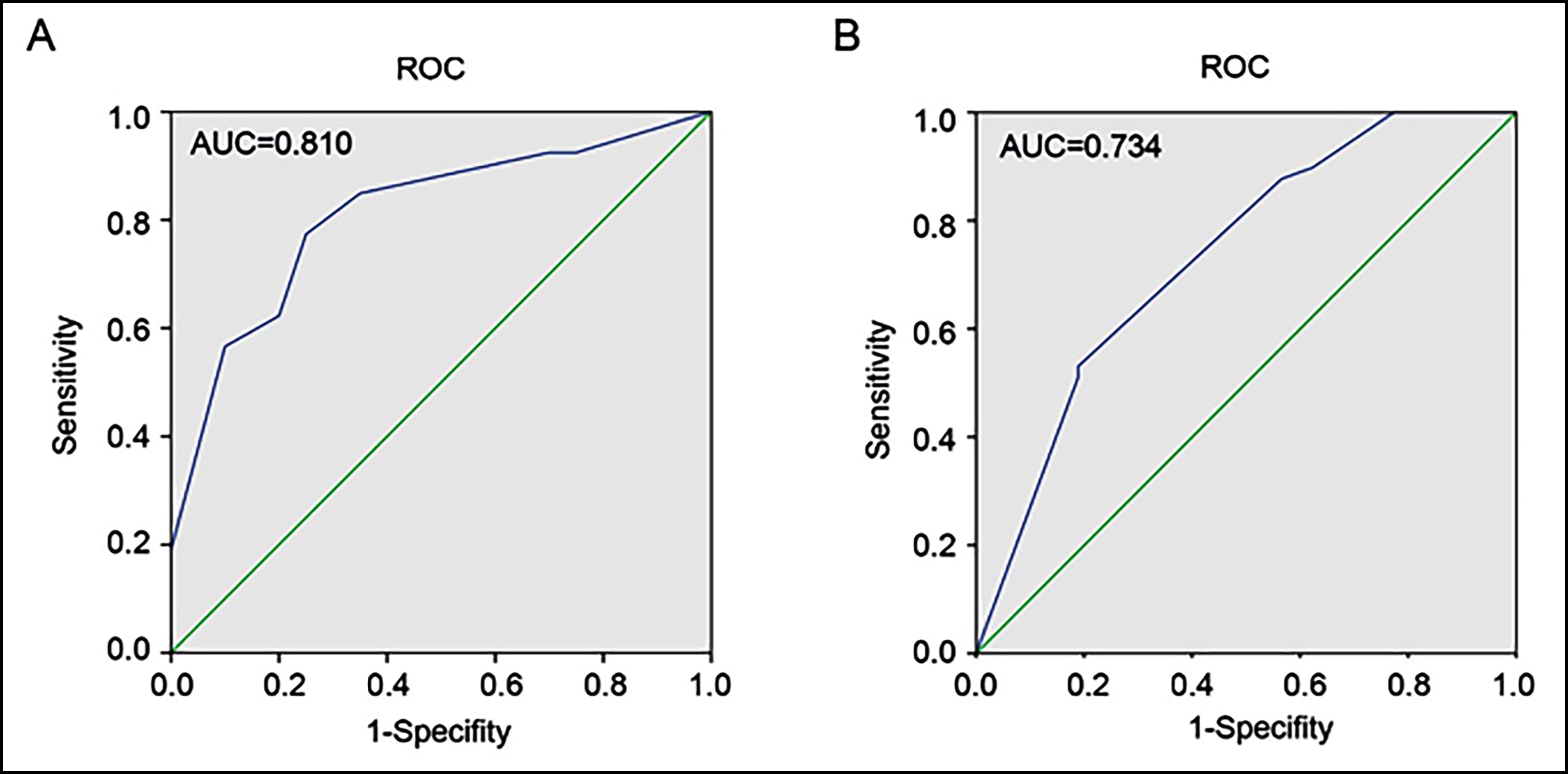 Figure 4: Analysing ROC and AUC of CBX2 in CRA and CRC. (A) ROC curve with corresponding AUC value for CBX2 when classifying CRA and mucosa. (B) Distinguish CRC from CRA through ROC curve with corresponding AUC value for CBX2.
Figure 4: Analysing ROC and AUC of CBX2 in CRA and CRC. (A) ROC curve with corresponding AUC value for CBX2 when classifying CRA and mucosa. (B) Distinguish CRC from CRA through ROC curve with corresponding AUC value for CBX2.
ROC: Receiver operating characteristic curve, AUC: Area under the curve.
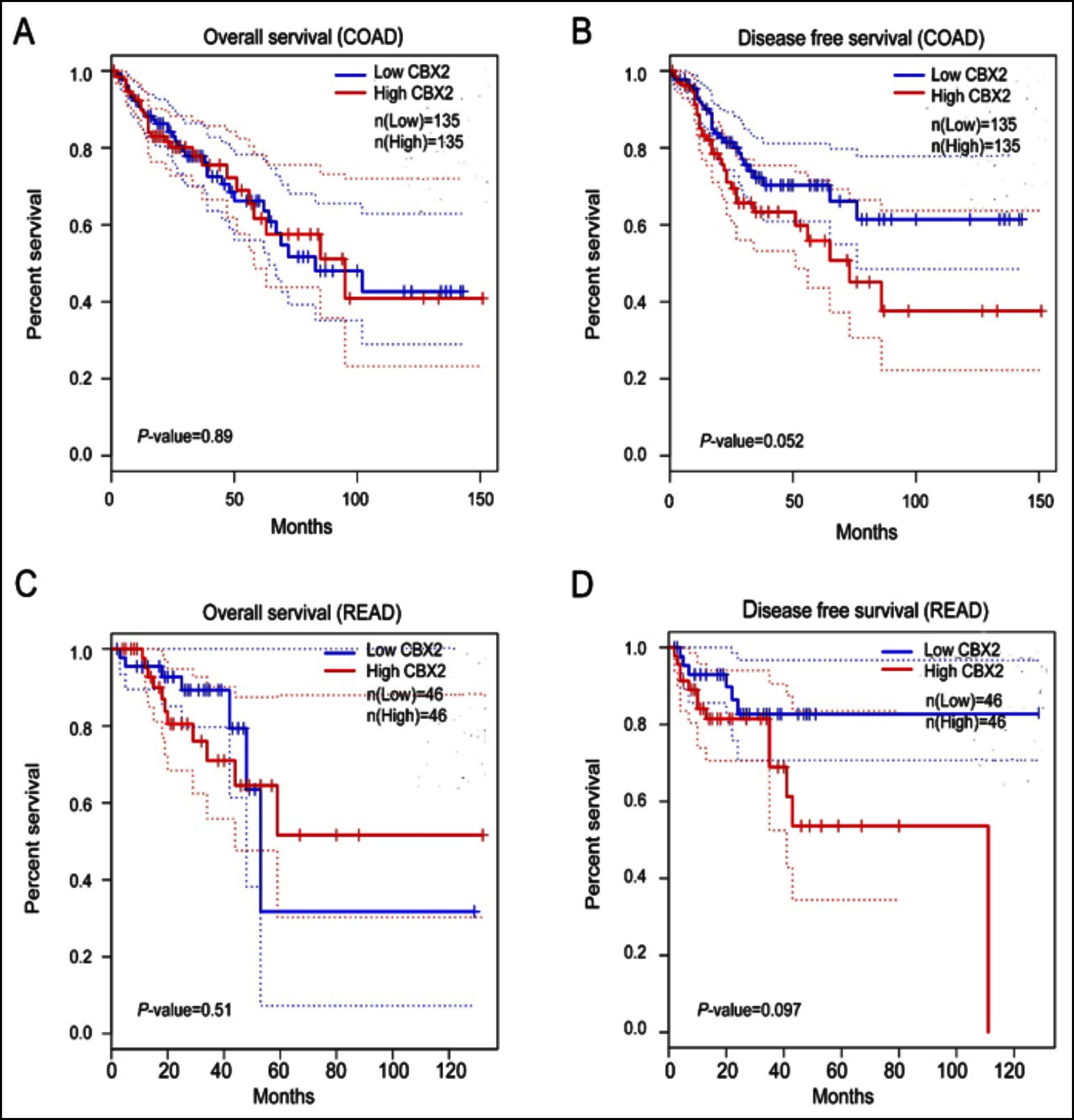 Figure 5: Kaplan–Meier survival curves for COAD and READ with high and low CBX2 mRNA expression in GEPIA. (A-B) Kaplan–Meier survival curves for overall survival and disease free survival in COAD according to CBX2 expression. (C-D) Kaplan–Meier survival curves for overall survival and disease free survival in READ according to CBX2 expression. COAD, colon adenocarcinoma; READ, rectum adenocarcinoma. High and low CBX2 expression levels are respectively indicated in red and blue.
Figure 5: Kaplan–Meier survival curves for COAD and READ with high and low CBX2 mRNA expression in GEPIA. (A-B) Kaplan–Meier survival curves for overall survival and disease free survival in COAD according to CBX2 expression. (C-D) Kaplan–Meier survival curves for overall survival and disease free survival in READ according to CBX2 expression. COAD, colon adenocarcinoma; READ, rectum adenocarcinoma. High and low CBX2 expression levels are respectively indicated in red and blue.
Furthermore, CBX2 immunohistochemical intensity was quantified. Among 122 specimens, 81 were high (staining score≥6) and 41 were low (staining score<6) for CBX2. CBX2 hyperexpression rate was then calculated according to these three histopathological categories (mucosa, adenoma and adenocarcinoma). The present result revealed that the ratio of high-level expression CBX2 increased in the following order: mucosa (20%, 4/20); adenoma (62.26%, 33/53) and adenocarcinoma (89.8%, 44/49) (Figure 3b). According to immunohistochemical score, the difference in the mean CBX2 level among mucosa (mean score±SD: 2.65±2.475), adenoma (mean score±SD: 6.604±3.626), and adenocarcinoma (mean score ± SD: 9.612 ± 2.702) was statistically significant (p <0.0001, Figure 3c). However, the expression of CBX2 was not significantly different between low-grade adenoma and high-grade adenoma (Table I). Altogether, those data indicated that the expression of CBX2 was gradually augmented during colorectal tumorigenesis, from normal mucosa to adenoma and adenocarcinoma. Then, ROC and AUC were utilised to predict the diagnostic value of CBX2 in distinguishing CRA from mucosa, and CRC from CRA (Figure 4). The area under the curve (AUC) values of CBX2 were 0.810 and 0.734, respectively.
Fifty-three patients with adenoma were analysed and the clinicopathological characteristics were shown in Table I. Table I also illustrated the correlation between CBX2 expression level and clinicopathological variables. However, no statistically significant correlations were observed between CBX2 expression level and parameters including age (p=0.748), gender (p=0.265), size (p=0.416), risk (p=0.416), or location (p=0.722).
Table I: Correlation of CBX2 with clinicopathological variables in patients with CRA.
|
Characteristics |
No of patients n=53 (%) |
CBX2 expression |
p-value |
|
|
low |
high |
|||
|
Age |
||||
|
<65 |
28 (52.8) |
10 |
18 |
0.748 |
|
≥65 |
25 (47.2) |
10 |
15 |
|
|
Gender |
||||
|
Male |
32 (60.4) |
14 |
18 |
0.265 |
|
Female |
21 (39.6) |
6 |
15 |
|
|
Histology |
||||
|
Low-risk adenoma |
28 (52.8) |
12 |
16 |
0.416 |
|
High-risk adenoma |
25 (47.2) |
8 |
17 |
|
|
Size |
||||
|
≤10mm |
28 (43.4) |
12 |
16 |
0.416 |
|
>10mm |
25 (37.7) |
8 |
17 |
|
|
Location |
||||
|
Right colon |
21 (39.6) |
7 |
14 |
0.722 |
|
Left colon |
26 (49.1) |
10 |
16 |
|
|
Rectum |
6 (11.3) |
3 |
3 |
|
Table II: Association of CBX2 with clinicopathological characteristics in CRC.
|
Characteristics |
No of patients n=49 (%) |
CBX2 expression |
p-value |
|
|
low |
high |
|||
|
Age |
||||
|
<60 |
20 (40.8) |
2 |
18 |
1 |
|
≥60 |
29 (59.2) |
3 |
26 |
|
|
Gender |
||||
|
Male |
31 (63.3) |
5 |
26 |
0.143 |
|
Female |
18 (36.7) |
0 |
18 |
|
|
BMI |
||||
|
<22 |
32 (65.3) |
5 |
27 |
0.149 |
|
≥22 |
17 (34.7) |
0 |
17 |
|
|
Diabetes |
||||
|
– |
39 (79.6) |
5 |
34 |
0.569 |
|
+ |
10 (20.4) |
0 |
10 |
|
|
Location |
||||
|
Right colon |
9 (18.4) |
1 |
8 |
0.831 |
|
Lift colon |
19 (38.8) |
1 |
18 |
|
|
Rectum |
21 (42.8) |
3 |
18 |
|
|
Lymphatic invasion |
||||
|
– |
25 (51) |
4 |
21 |
0.349 |
|
+ |
24 (49) |
1 |
23 |
|
|
Metastasis |
||||
|
– |
29 (59.2) |
2 |
27 |
0.636 |
|
+ |
20 (40.8) |
3 |
17 |
|
|
Stage |
||||
|
ⅠⅡ |
17 (34.5) |
2 |
15 |
1 |
|
ⅢⅣ |
32 (65.5) |
3 |
29 |
|
The correlation between CBX2 expression and clinicopathological factors of CRC was evaluated and shown in Table II. CBX2 expression did not vary significantly in terms of gender (p=1.00), age (<60 years versus ≥60 years, p=0.143), segment of large intestine (right, left and rectum, p=0.831), stage (I, II versus III, IV, p=1.00), and BMI (<22 versus ≥22, p=0.149). Moreover, CBX2 expression did not correlate with diabetes (p=0.569), metastasis (p=0.387) or lymphatic invasion (p=0.349).
The analyses of OS and DFS were performed via GEPIA (http://gepia.cancer-pku.cn/) using colorectal adenocarcinoma samples from TCGA data).
The relationship between the clinical outcome of CRC patients and the mRNA expression of CBX2 was evaluated by Kaplan-Meier survival analysis and log-rank test. Analysis was performed via GEPIA using colorectal adenocarcinoma samples from TCGA data, including 135 COAD patients and 46 READ patients. Kaplan-Meier survival curves indicated that there was no correlation between high CBX2 expression and short OS (overall survival) in COAD (log-rank test, p=0.89) and READ (log-rank test, p=0.51) (Figure 5a and 5c). The DFS (disease-free survival) of patients with high CBX2 patients is shorter than those with low expression, but being not obviously significant in COAD (log-rank test, p=0.052) and READ (log-rank test, p=0.097, Figure 5b and 5d).
DISCUSSION
Studying the evolutionary processes of colorectal adenomas, including its progression to cancer, remains challenging because adenomas are often removed when detected during colonoscopy.20 However, there is a practical need to understand the adenoma biology and its progress to CRC. The selection of the most beneficial treatment regimens in CRC remains challenging due to lack of prognostic markers. In this study, the authors have focused on identifying novel biomarker of sporadic CRA and CRC to characterise these neoplasms.
Recently, studies about DEGs or molecular biomarkers of CRA and CRC have been increasingly reported. Based on these results, aberrantly significant DEGs and pathways in CRC have been identified. For example, CXCL3, and DSN1 are shown to have diagnostic value with respect to the occurrence of colorectal cancer.21,22
CBX2 is an important member of the CBX protein family and a part of the PRC1 complex involved in regulating chromatin structure.8 CBX2 is widely considered to be a tumor‑promoting gene and plays an important role in the occurrence and development of many tumors, such as pancreatic ductal adenocarcinoma and esophageal cancer.23,24 PCAT6 could upregulate the expression of oncogene CBX2 to promote tumorigenesis and progression of pancreatic ductal adenocarcinoma.23 Similarly, high CBX2 expression was significantly related with shorter disease-specific survival, hematogenous recurrence, and overall recurrence in esophageal cancer.24 However, the expression and function of CBX2 is not reported in colorectal cancer.
In the present study, it was found that CBX2 was highly expressed in CRA (GSE8671) and CRC (GEPIA) compared to normal mucosa. The expression of CBX2 was successfully validated by immunohistochemical staining among normal mucosa, adenoma and adenocarcinoma. Semi-quantitative analysis of immunohistochemical staining showed CBX2 expression was highest in adenocarcinoma, followed by adenoma, and the lowest level in mucosa. As an oncogene, CBX2 hyperexpression may be regulated by methylation. The authors employed the Ualcan, and demonstrate the promoter hypomethylation level of CBX2. CBX2 expression in adenoma and adenocarcinoma had no significant correlation with clinicopathological factors, such as age, gender and location. Besides, high expression of CBX2 in adenocarcinoma showed no obvious association with tumor metastasis and stage. The authors did not detect a significant correlation between CBX2 expression and the overall survival of patients with CRC. Although CBX2 expression had a negative correlation with the patient's disease-free survival, there was no obvious significance.
Different from previous reports on CBX2 in other tumors, this study suggested that the expression of CBX2 increased along the sequence of mucosa-adenoma-adenocarcinoma, but there was no prognostic significance of CBX2 in CRC. High expression of CBX2 increased through the colorectal mucosa-adenoma-adenocarcinoma sequence indicating it may promote the transition of mucosal epithelium to adenoma, or even into adenocarcinoma.
There are still several limitations in this study. The number of specimens evaluated was relatively small for this retrospective study. The function of CBX2 in colorectal neoplasm needs to be confirmed through more assays, and the mechanism underlying the occurrence of CBX2 in CRA and CRC remains to be elucidated.
CONCLUSION
CBX2 expression is gradually augmented along the mucosa–adenoma–adenocarcinoma sequence, which may define malignant potential of adenoma evolving into adenocarcinoma. Besides, CBX2 might also serve as a novel clinical diagnostic marker and potential prevention and treatment target for CRC.
FUNDING:
This work was supported by the grant from National Natural Science Foundation of China (81630016).
ETHICAL APPROVAL:
Ethical approval was obtained for this study from the Ethical Committee of Medical Research, Huashan Hospital of Fudan University, Shanghai, China. (2017M-003).
PATIENTS' CONSENT:
Informed consents were obtained from all patients to publish the data concerning this study.
CONFLICT OF INTEREST:
The authors declared no conflict of interest.
AUTHORS’ CONTRIBUTION:
BW, FL: Designed the experiments.
BW, XW: Performed the experiments, analysed the data.
FL, JL: Supervised the work.
YT, BW, MH: Wrote the manuscript.
REFERENCES
- Siskova A, Cervena K, Kral J, Hucl T, Vodicka P, Vymetalkova V. Colorectal adenomas-genetics and searching for new molecular screening biomarkers. Int J Mol Sci 2020; 21(9):3260. doi: 10.3390/ijms21093260.
- Castellsagué E, Rivera B, Foulkes W. Colorectal Adenomas. New Eng J Med 2016; 375: 389. doi: 10.1056/NEJMc160 4867.
- Aceto GM, Catalano T, Curia MC. Molecular aspects of colorectal adenomas: The interplay among micro-environment, oxidative stress, and predisposition. Biomed Res Int 2020; 2020: 1726309. doi: 10.1155/2020/1726309.
- Jones S, Chen W, Parmigiani G, Diehl F, Beerenwinkel N, Antal T, et al. Comparative lesion sequencing provides insights into tumor evolution. Proc Natl Acad Sc USA 2008; 105(11):4283-8. doi: 10.1073/pnas.0712345105.
- Soetikno RM, Kaltenbach T, Rouse RV, Park W, Maheshwari A, Sato T, et al. Prevalence of nonpolypoid (flat and depressed) colorectal neoplasms in asymptomatic and symptomatic adults. JAMA 2008; 299(9):1027-35. doi: 10.1001/jama.299.9.1027.
- Luo Y, Wong CJ, Kaz AM, Dzieciatkowski S, Carter KT, Morris SM, et al. Differences in DNA methylation signatures reveal multiple pathways of progression from adenoma to colorectal cancer. Gastroenterol 2014; 147(2):418-29.e8. doi: 10.1053/j.gastro.2014.04.039.
- Hisabe T, Hirai F, Matsui T. Development and progression of colorectal cancer based on follow-up analysis. Dig Endosc 2014; 26(2):73-7. doi: 10.1111/den.12276.
- Baumann C, Zhang X, La Fuente RD. Loss of CBX2 induces genome instability and senescence-associated chromo-somal rearrangements. J Cell Biol 2020; 219(11):e201910149. doi: 10.1083/jcb.201910149.
- Simon J, Kingston R. Occupying chromatin: Polycomb mechanisms for getting to genomic targets, stopping transcriptional traffic, and staying put. Mol Cell 2013; 49(5):808-24. doi: 10.1016/j.molcel.2013.02.013.
- Margueron R, Reinberg D. The Polycomb complex PRC2 and its mark in life. Nature 2011; 469(7330):343-9. doi: 10.1038/nature09784.
- Pereira C, Piccolo F, Tsubouchi T, Sauer S, Ryan N, Bruno L, et al. ESCs require PRC2 to direct the successful reprogramming of differentiated cells toward pluripotency. Cell Stem Cell 2010; 6(6):547-56. doi: 10.1016/j.stem. 2010.04.013.
- Morey L, Pascual G, Cozzuto L, Roma G, Wutz A, Benitah S. et al. Nonoverlapping functions of the polycomb group Cbx family of proteins in embryonic stem cells. Cell Stem Cell 2012; 10(1):47-62. doi: 10.1016/j.stem.2011.12.006.
- Sun D, Cao X, Wang C. Polycomb chromobox Cbx2 enhances antiviral innate immunity by promoting Jmjd3-mediated demethylation of H3K27 at the Ifnb promoter. Protein Cell 2019; 10(4):285-94. doi: 10.1007/s13238- 018-0581-0.
- Gu X, Wang X, Su D, Su X, Lin L, Li S, et al. CBX2 Inhibits neurite development by regulating neuron-specific genes expression. Front Mole Neurosci 2018; 11:46.doi: 10.3389/fnmol.2018.00046.
- Klauke K, Radulović V, Broekhuis M, Weersing E, Zwart E, Olthof S, et al. Polycomb Cbx family members mediate the balance between haematopoietic stem cell self-renewal and differentiation. Nature Cell Biol 2013; 15(4):353-62. doi: 10.1038/ncb2701.
- Mao J, Tian Y, Wang C, Jiang K, Li R, Yao Y, et al. CBX2 regulates proliferation and apoptosis via the phosphorylation of YAP in Hepatocellular Carcinoma. J Can 2019; 10(12):2706-19. doi: 10.7150/jca.31845.
- Zheng S, Lv P, Su J, Miao K, Xu H, Li M. Overexpression of CBX2 in breast cancer promotes tumor progression through the PI3K/AKT signaling pathway. Am J Trans Res 2019; 11(3):1668-82.
- Sun G, Li Y, Peng Y, Lu D, Zhang F, Cui X, et al. Identification of differentially expressed genes and biological characteristics of colorectal cancer by integrated bioinformatics analysis. J Cell Physiol 2019. doi: 10.1002/ jcp.28163.
- Liu XF, Li XY, Zheng PS, Yang WT. DAX1 promotes cervical cancer cell growth and tumorigenicity through activation of Wnt/beta-catenin pathway via GSK3beta. Cell Death Dis 2018; 9(3):339. doi: 10.1038/s41419-018-0359-6.
- Komor M, de Wit M, van den Berg J, Martens de Kemp S, Delis-van Diemen P, Bolijn A, et al. Molecular characterisation of colorectal adenomas reveals POFUT1 as a candidate driver of tumor progression. Int J Can 2020; 146(7):1979-92. doi: 10.1002/ijc.32627.
- Zhao Q, Jiang C, Gao Q, Zhang Y, Wang G, Chen X, et al. Gene expression and methylation profiles identified CXCL3 and CXCL8 as key genes for diagnosis and prognosis of colon adenocarcinoma. J Cell Physiol 2020; 235(9): 4902-12. doi: 10.1002/jcp.29368.
- Zhou H, Yang Z, Yue J, Chen Y, Chen T, Mu T, et al. Identification of potential hub genes via bioinformatics analysis combined with experimental verification in colorectal cancer. Mol Carcinog 2020; 59(4):425-38. doi: 10.1002/mc.23165.
- Wang W, Li X, Guan C, Hu Z, Zhao Y, Li W, et al. LncRNA PCAT6 promotes the proliferation, migration and invasion of pancreatic ductal adenocarcinoma via regulating miR-185-5p/CBX2 axis. Pathol Res Prac 2020; 216(9): 153074. doi: 10.1016/j.prp.2020.153074.
- Ueda S, Kanda M, Sato Y, Baba H, Nakamura S, Sawaki K, et al. Chromobox 2 expression predicts prognosis after curative resection of oesophageal squamous cell carcinoma. Cancer Genomics Proteomics 2020; 17(4): 391-400. doi: 10.21873/cgp.20198.